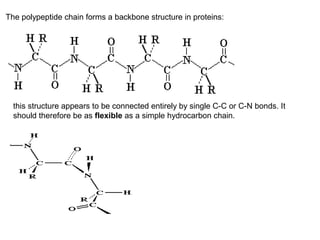
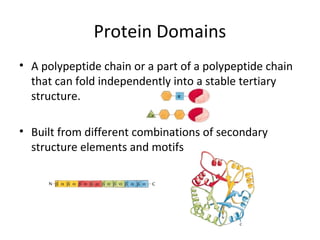
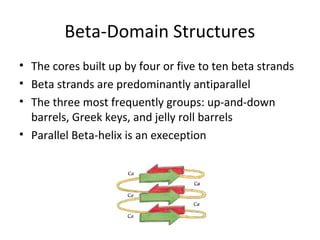
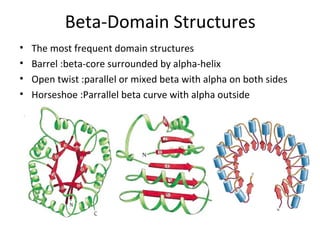
The document discusses new research that provides the most detailed view yet of tau protein structures found in Alzheimer's disease. Scientists from the UK and Indiana University were able to present high-resolution structures of tau filaments from an Alzheimer's patient's brain. These findings represent a major discovery and will help design new therapeutic agents by providing insight into tau's role in disease progression at the atomic level.