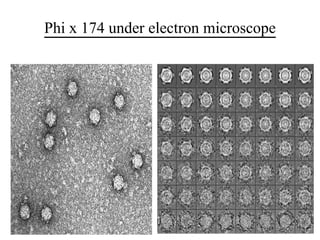
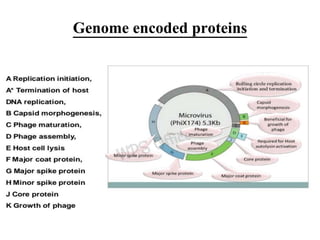
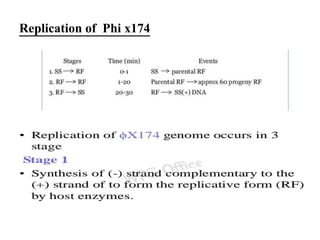
This document discusses the bacterial virus Phi X 174 phage. It begins with an introduction to bacteriophages and their discovery. It then provides the taxonomic classification of Phi X 174, placing it in the family Microviridae. The document outlines the history of Phi X 174, including its early study and sequencing. It describes the morphology and genome organization of Phi X 174, including its circular single-stranded DNA genome of 5386 nucleotides that encodes 10 genes and 11 proteins. The life cycle of Phi X 174 is summarized, from attachment and entry, to replication, assembly of mature virions, and release through cell lysis.