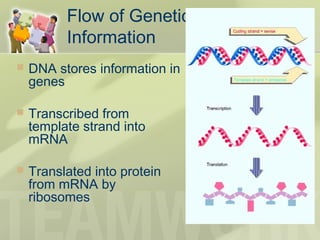
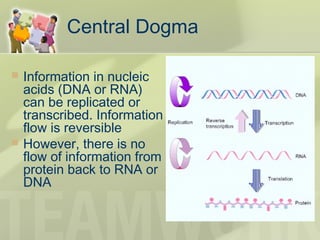
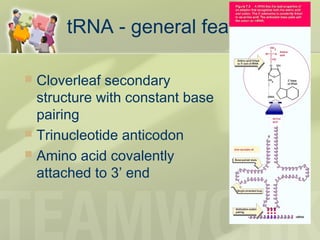
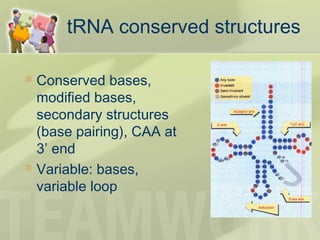
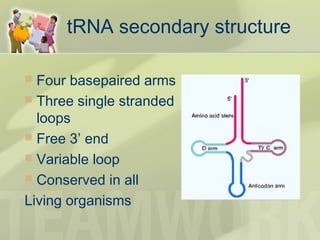
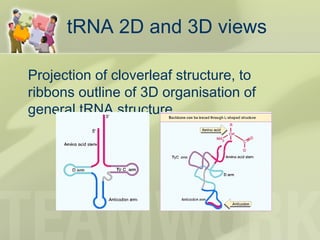
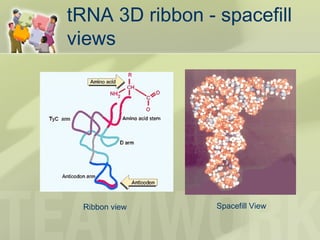
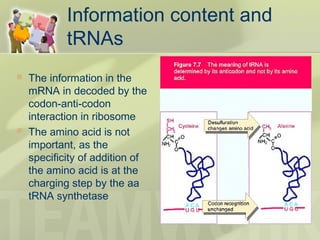
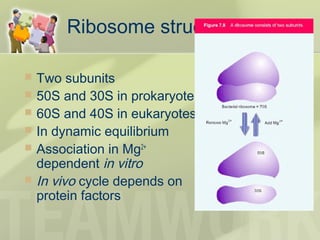
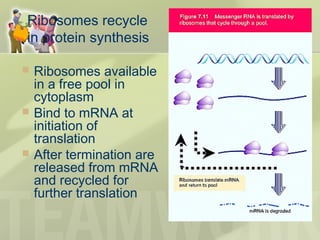
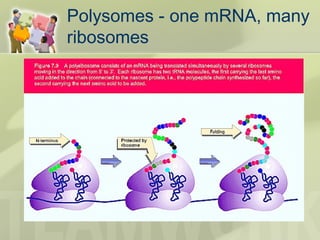
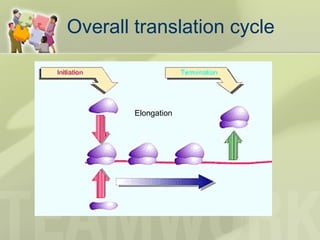
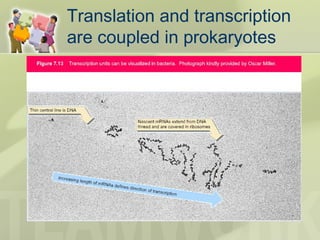
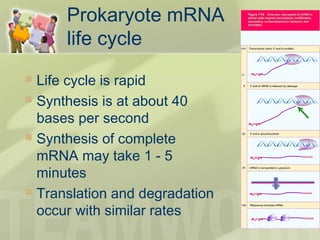
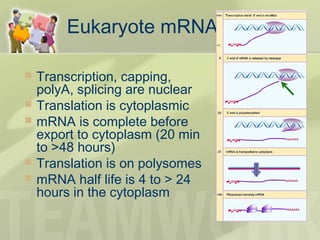
The document provides a detailed overview of prokaryote and eukaryote gene expression, focusing on the central dogma of molecular biology, which describes the flow of genetic information from DNA to RNA to protein. It explains key processes such as replication, transcription, and translation, while highlighting differences in mRNA structure and regulation between prokaryotes and eukaryotes. Additionally, the document outlines the roles and characteristics of tRNA and ribosomes in protein synthesis.