1. Gene expression can be regulated positively or negatively at the levels of transcription, RNA processing, translation and protein activity through the actions of regulatory proteins and hormones.
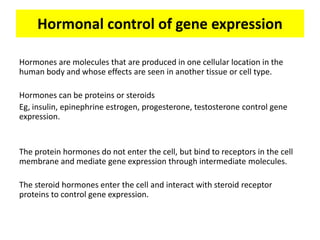
2. Hormones like steroids enter cells and bind nuclear receptors to activate transcription, while peptide hormones signal through cell surface receptors and secondary messengers.
3. Key mechanisms of transcriptional control include chromatin remodeling, DNA methylation, and the binding of transcription factors to regulatory sequences which can either promote or block transcription initiation.