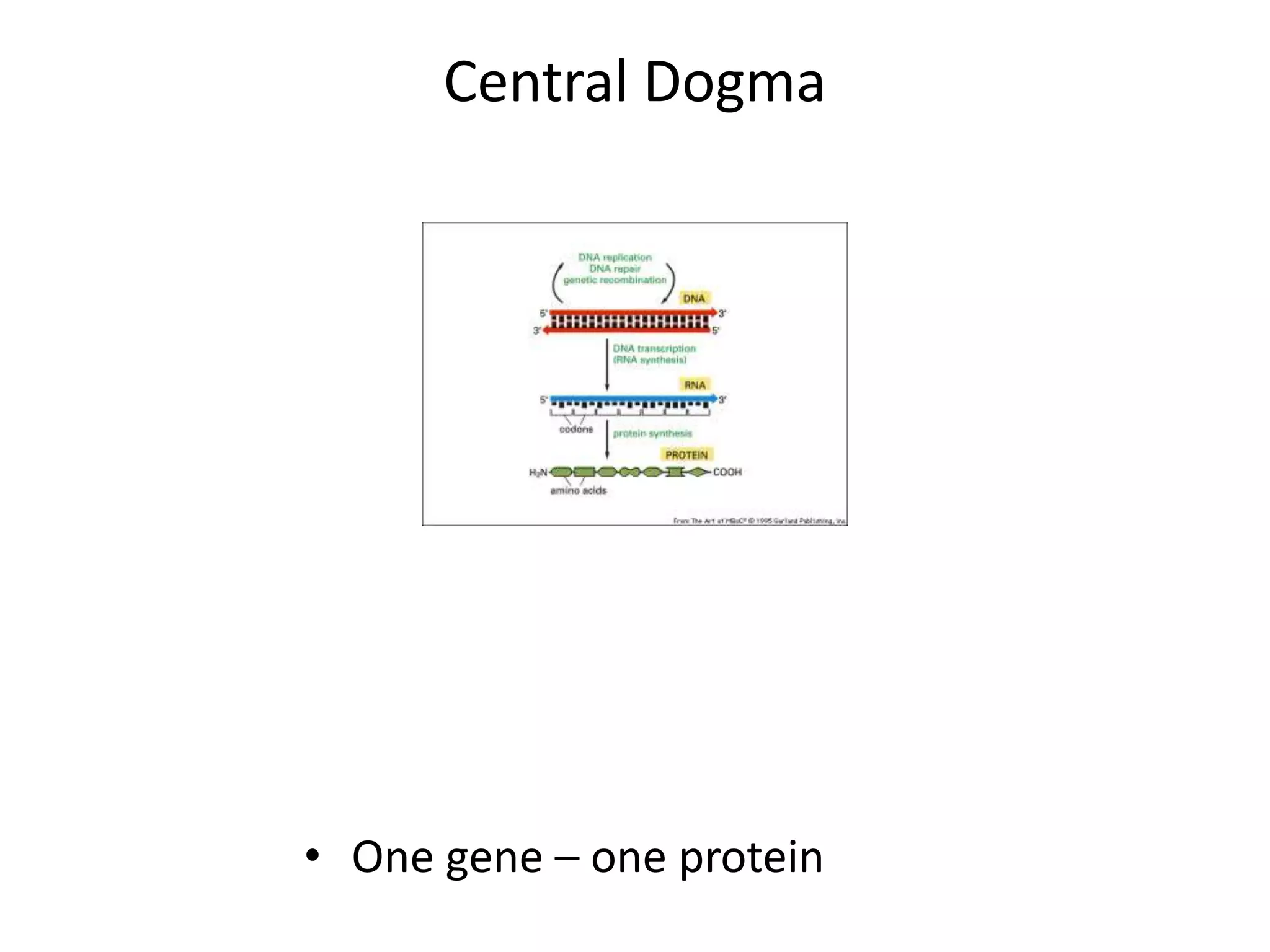
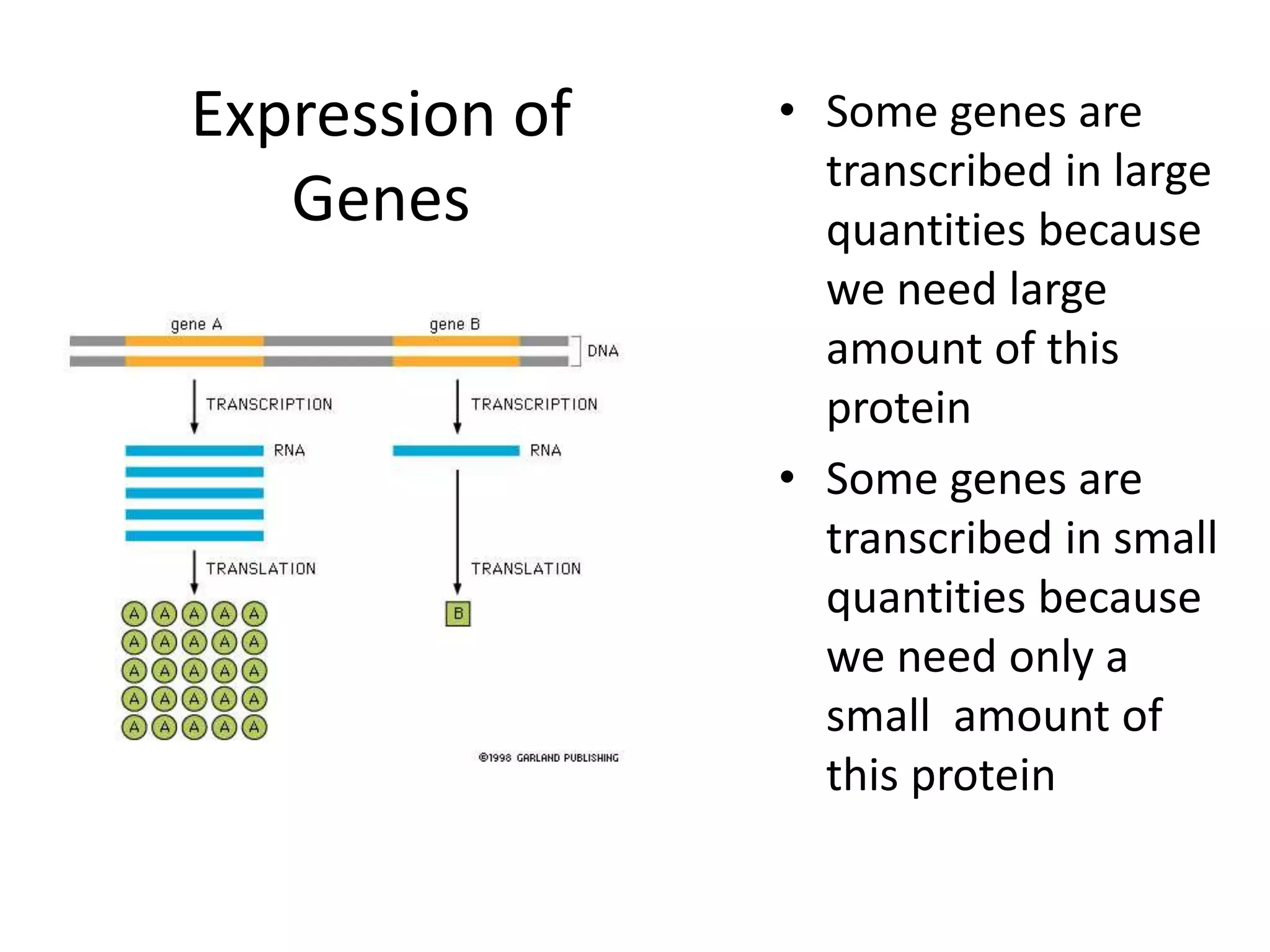
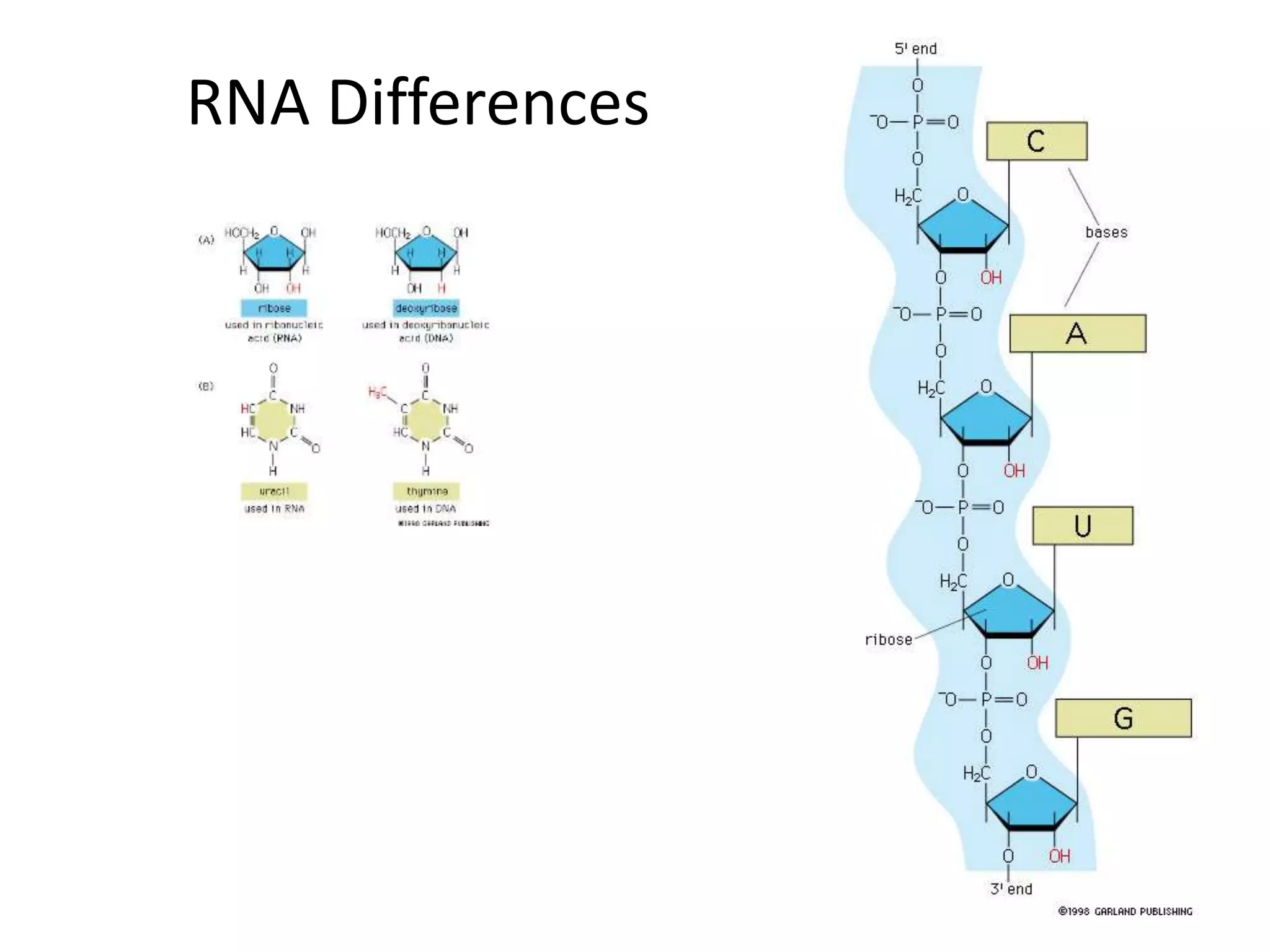
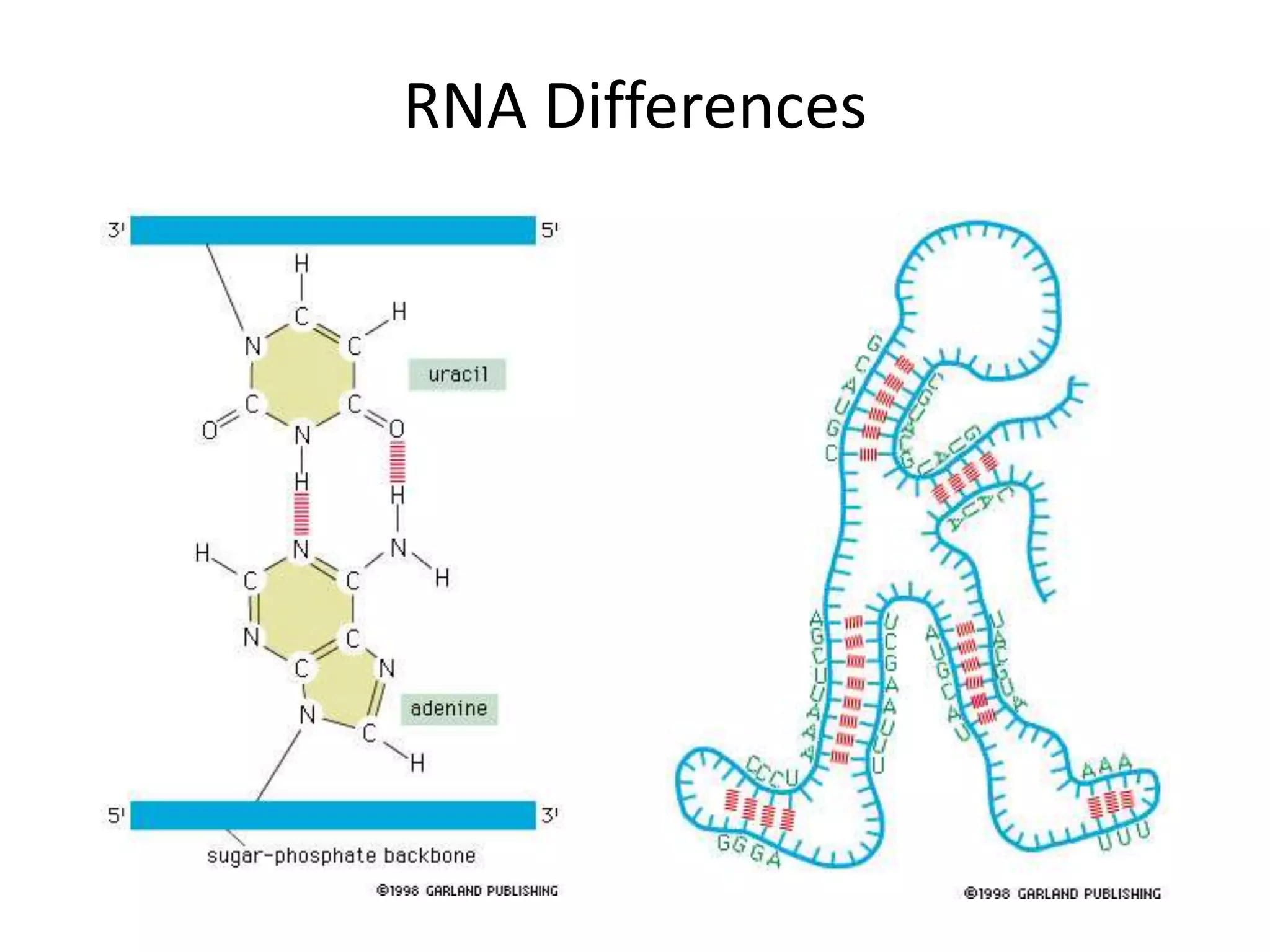
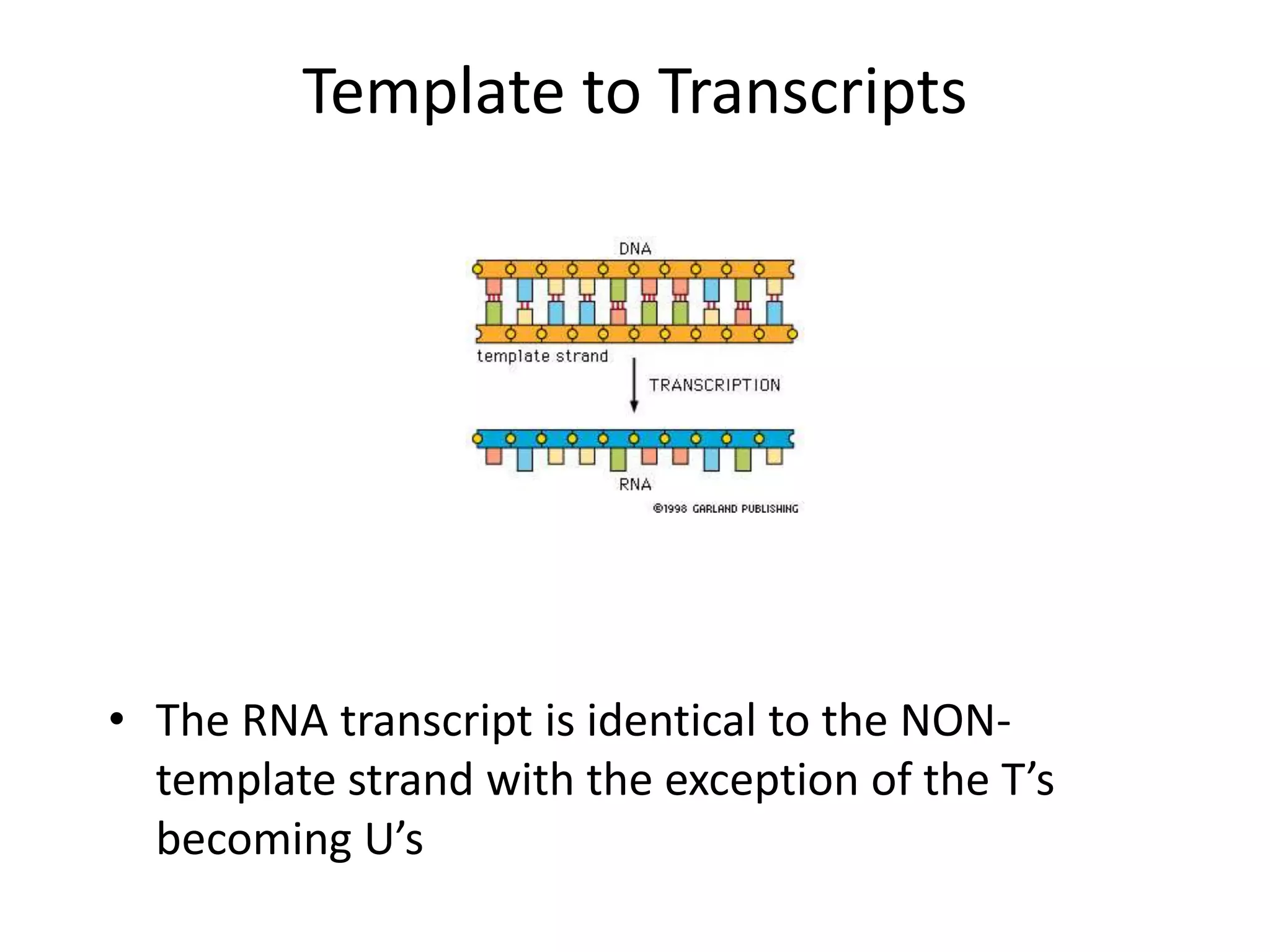
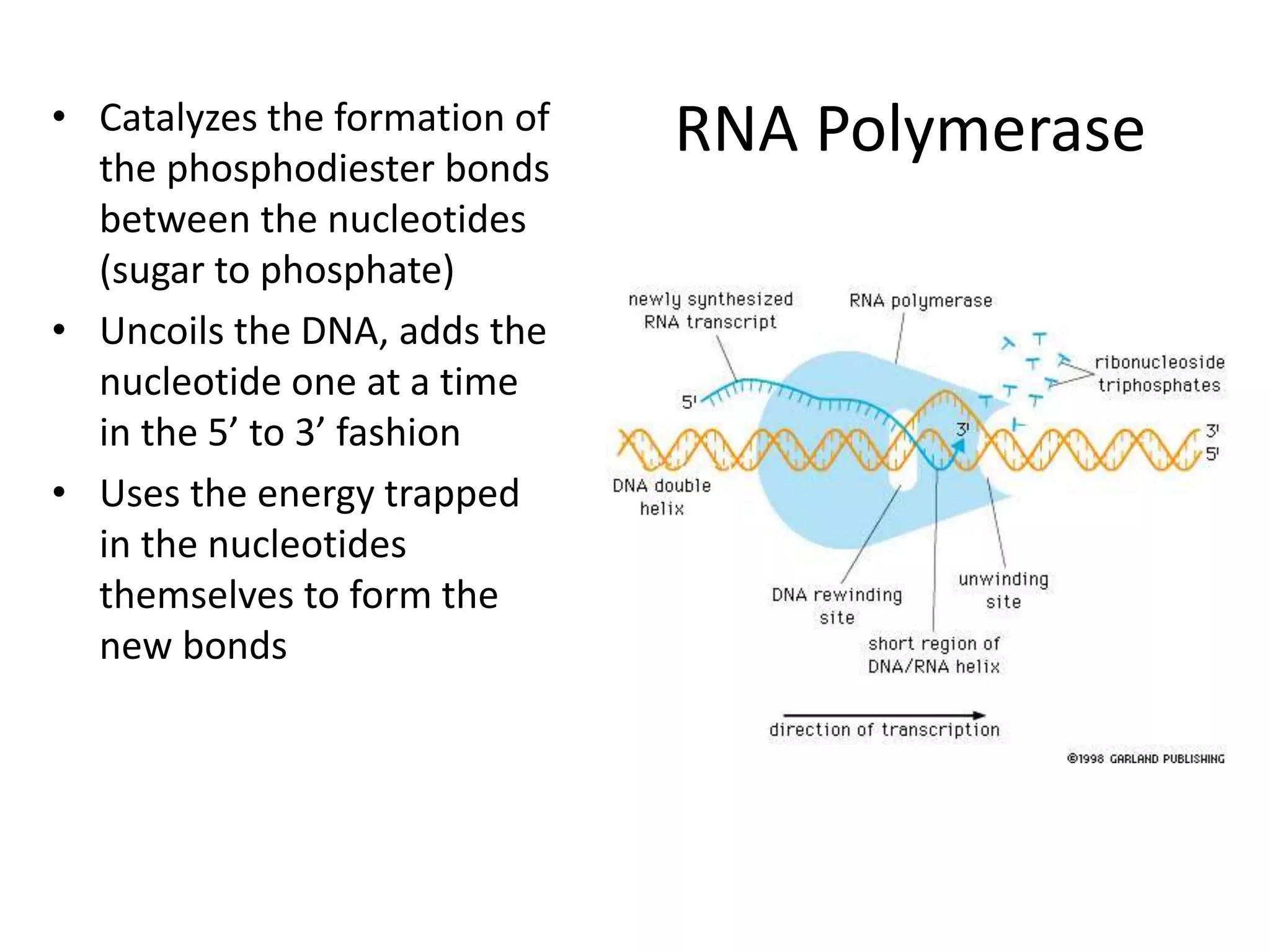
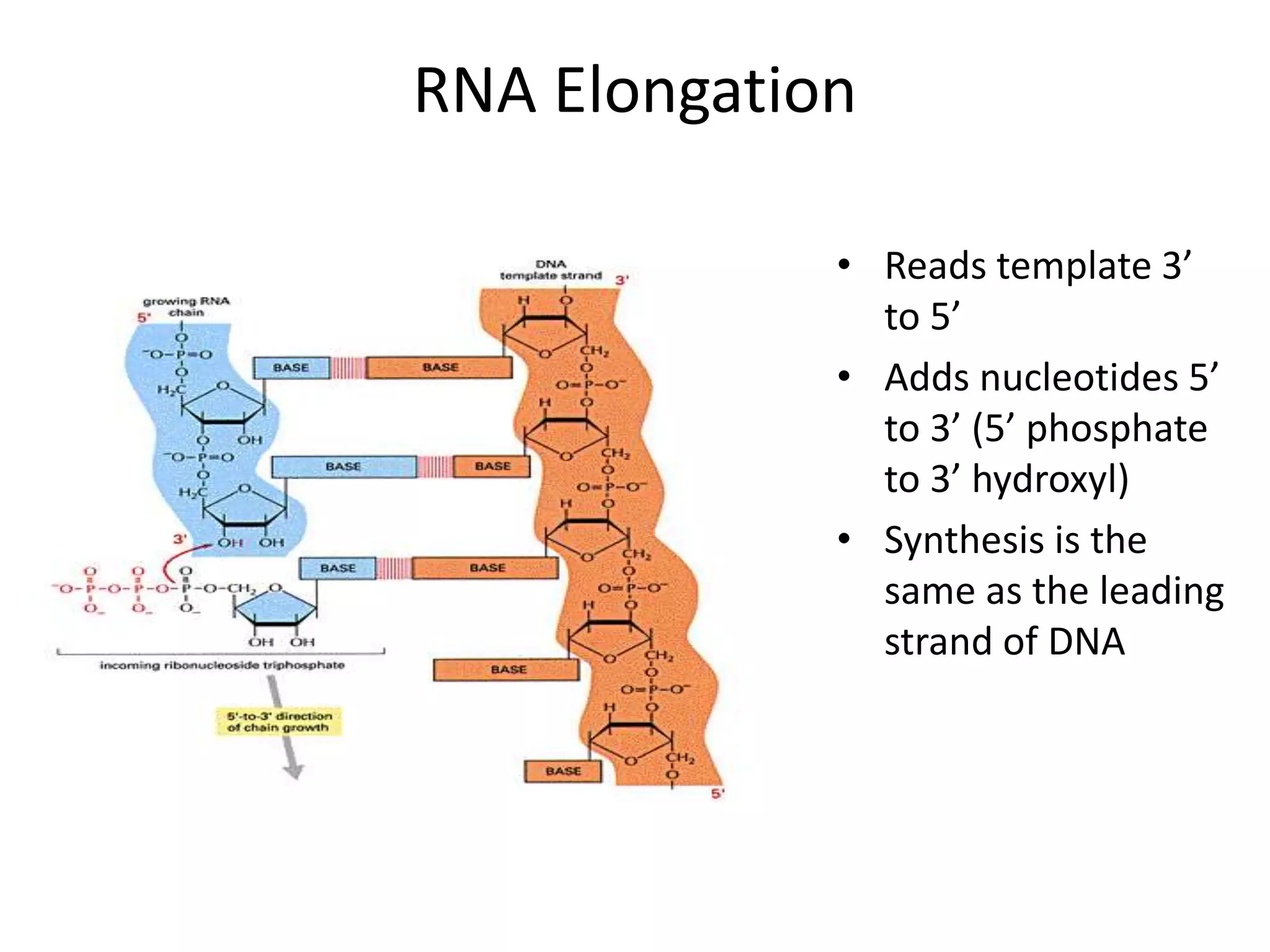
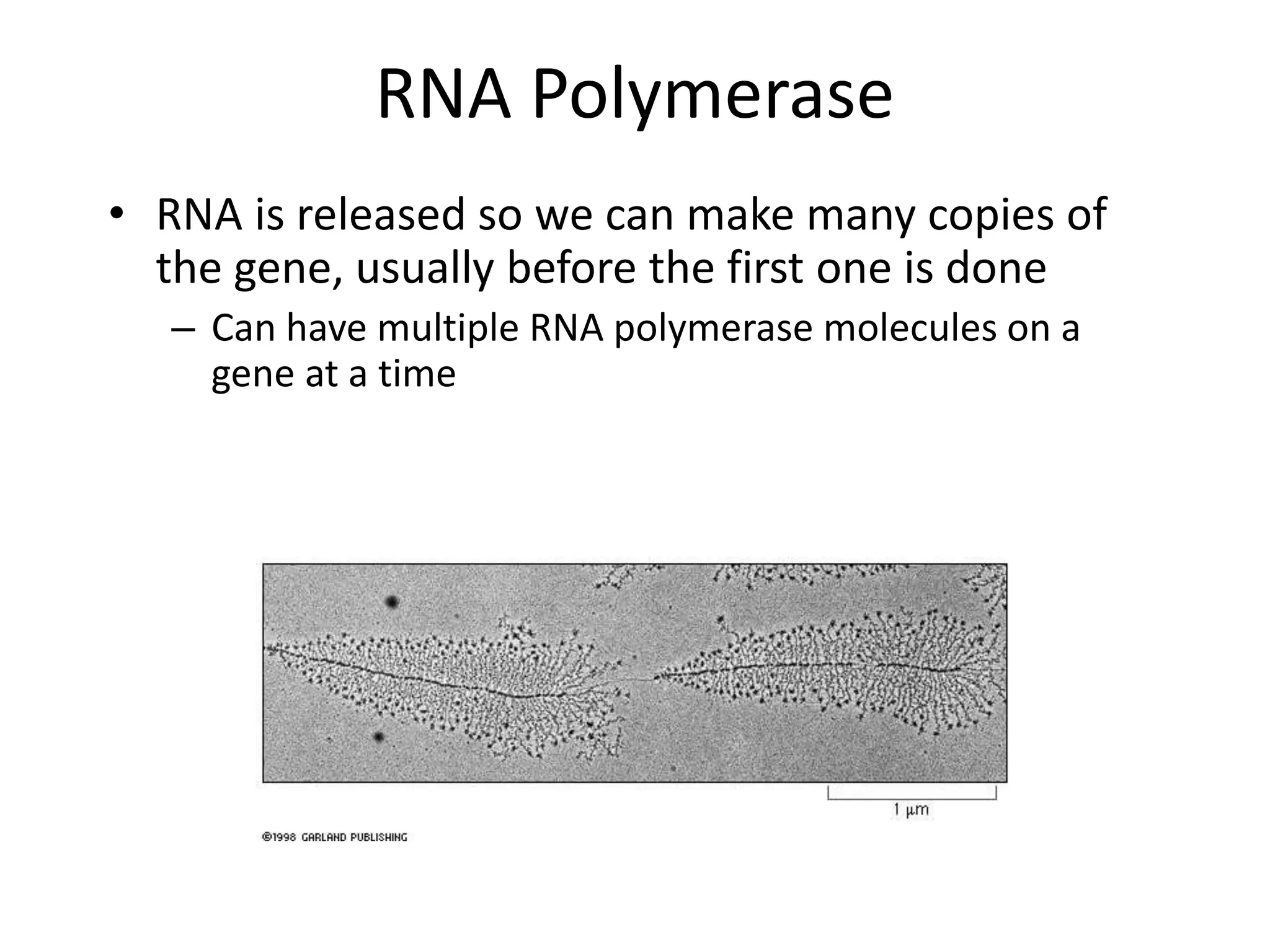
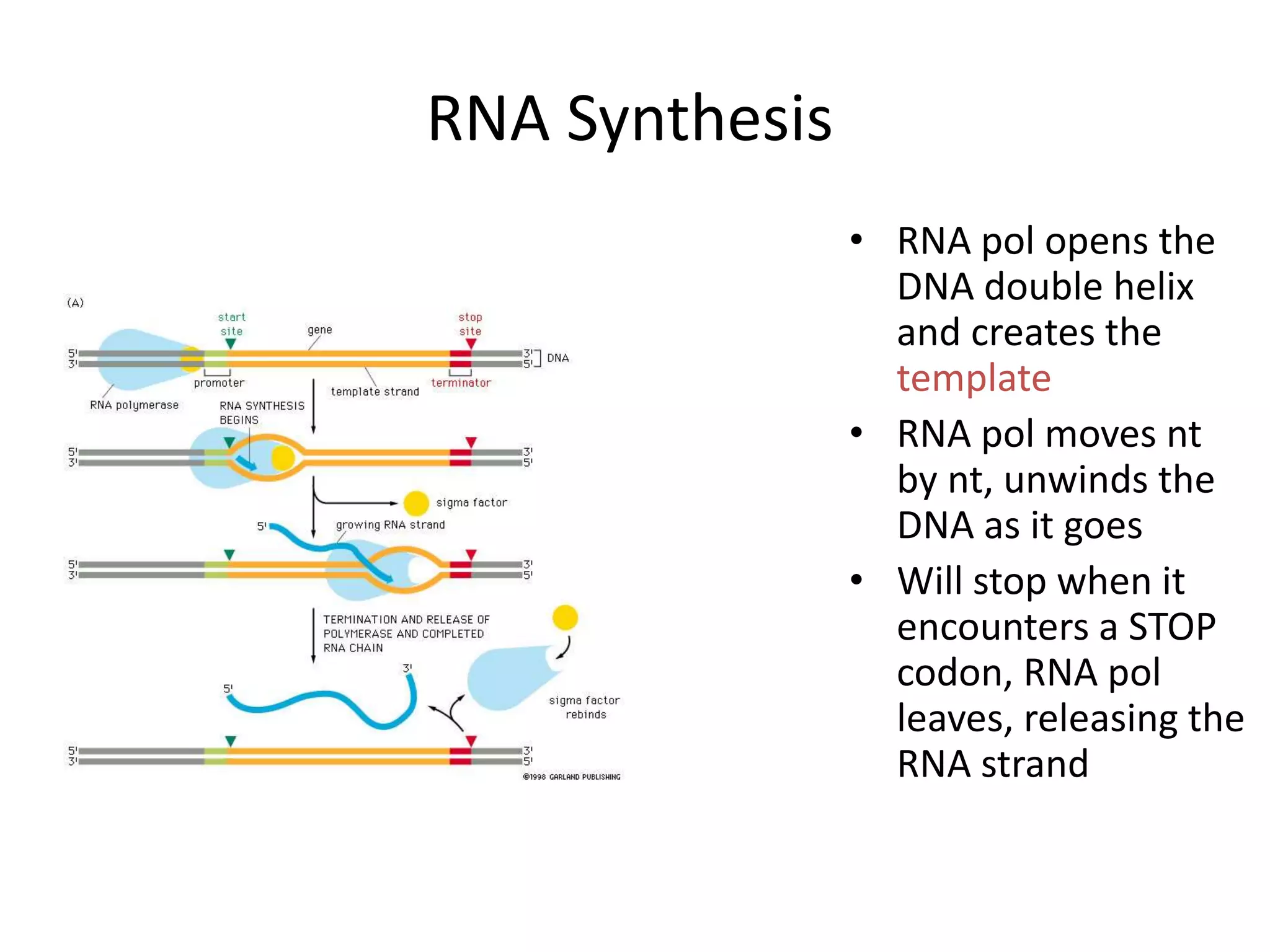
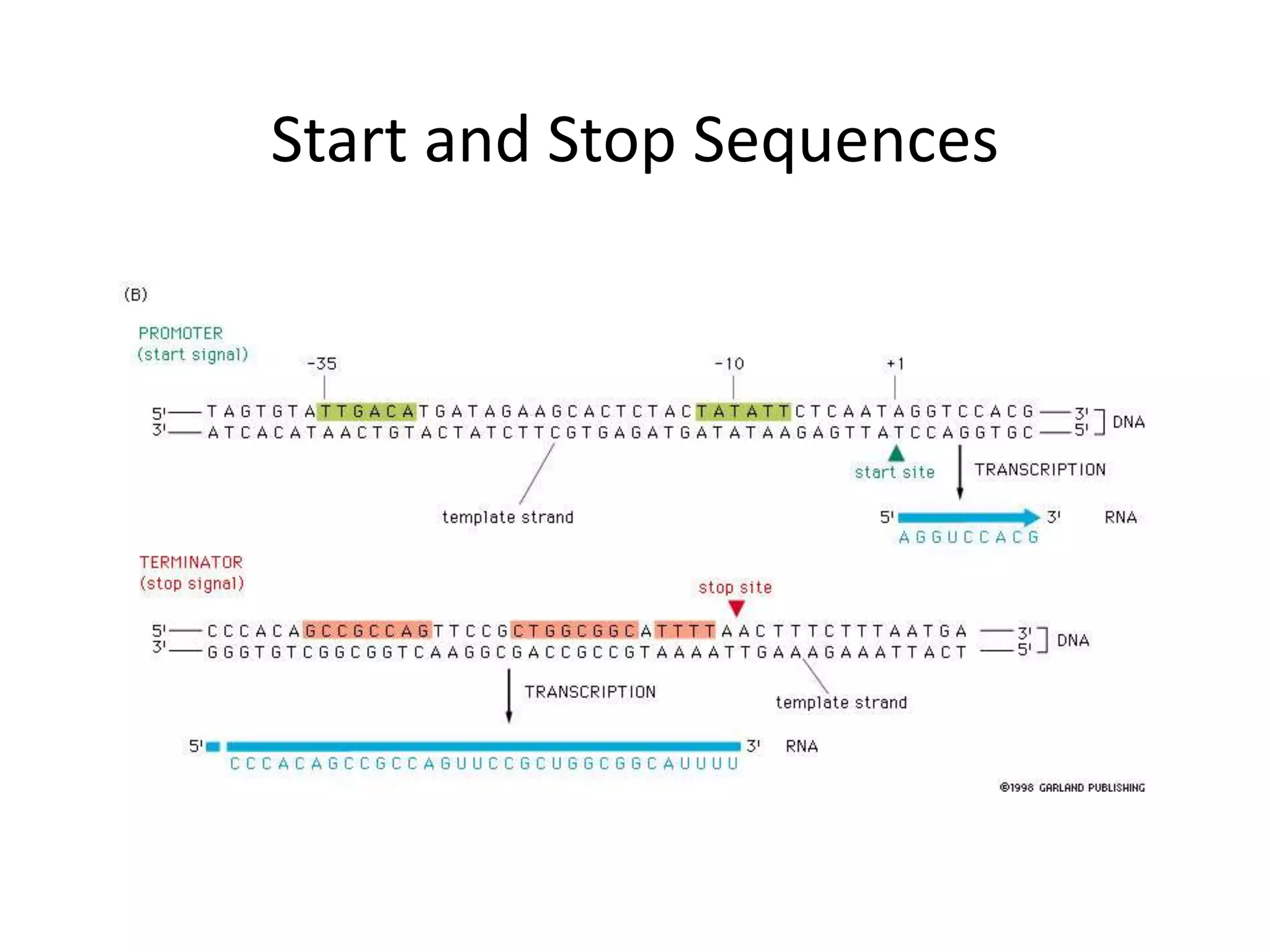
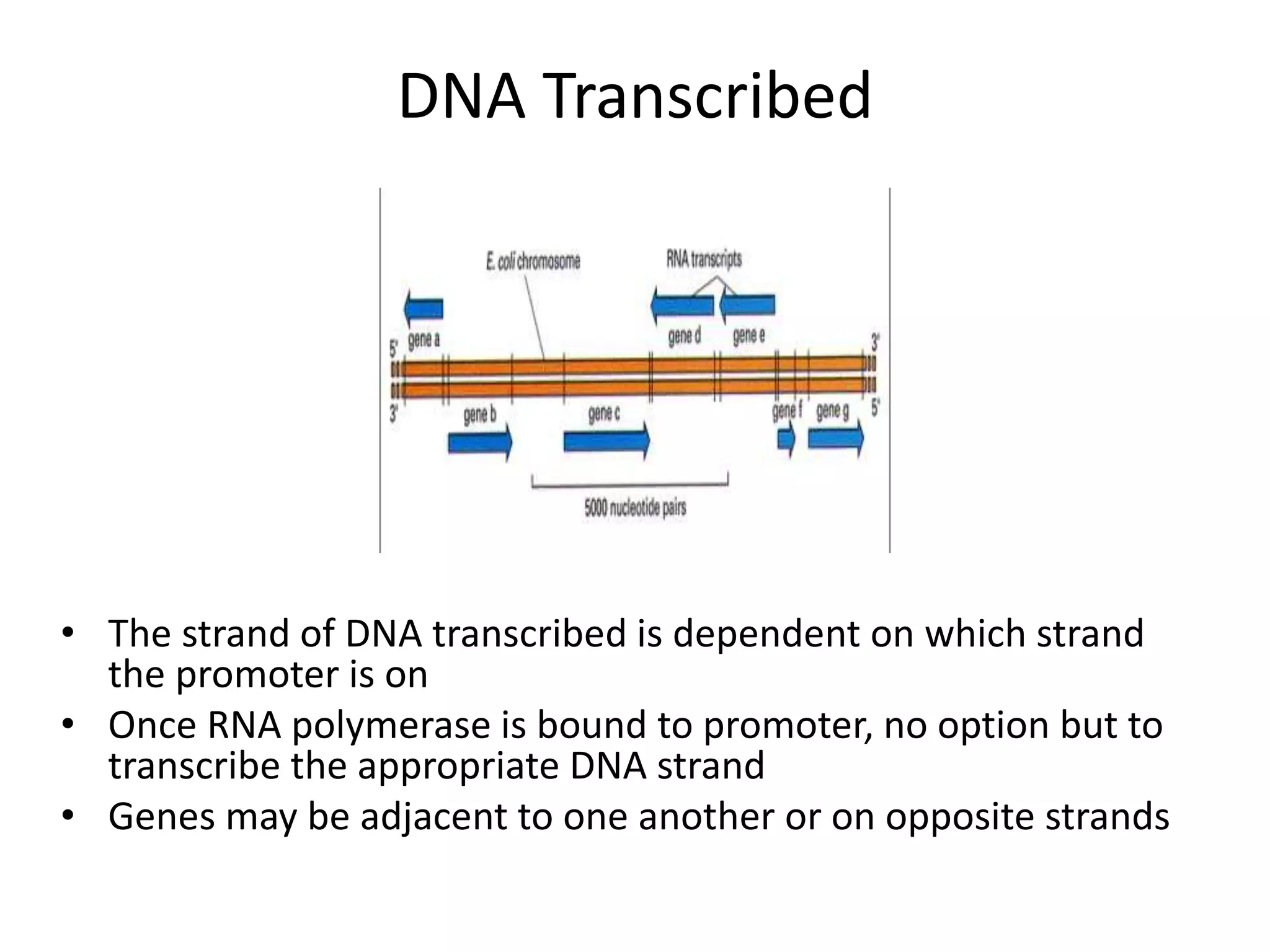
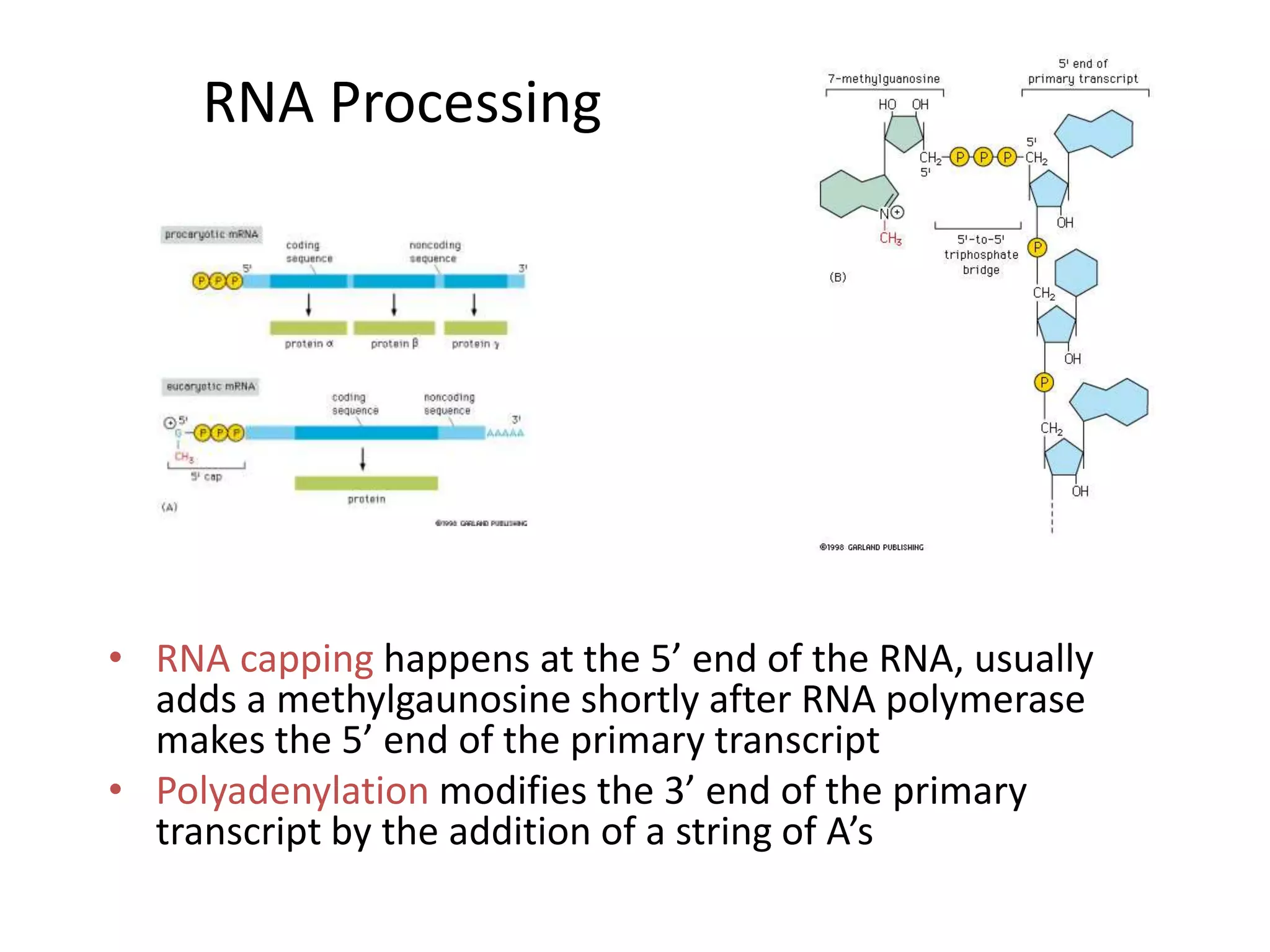
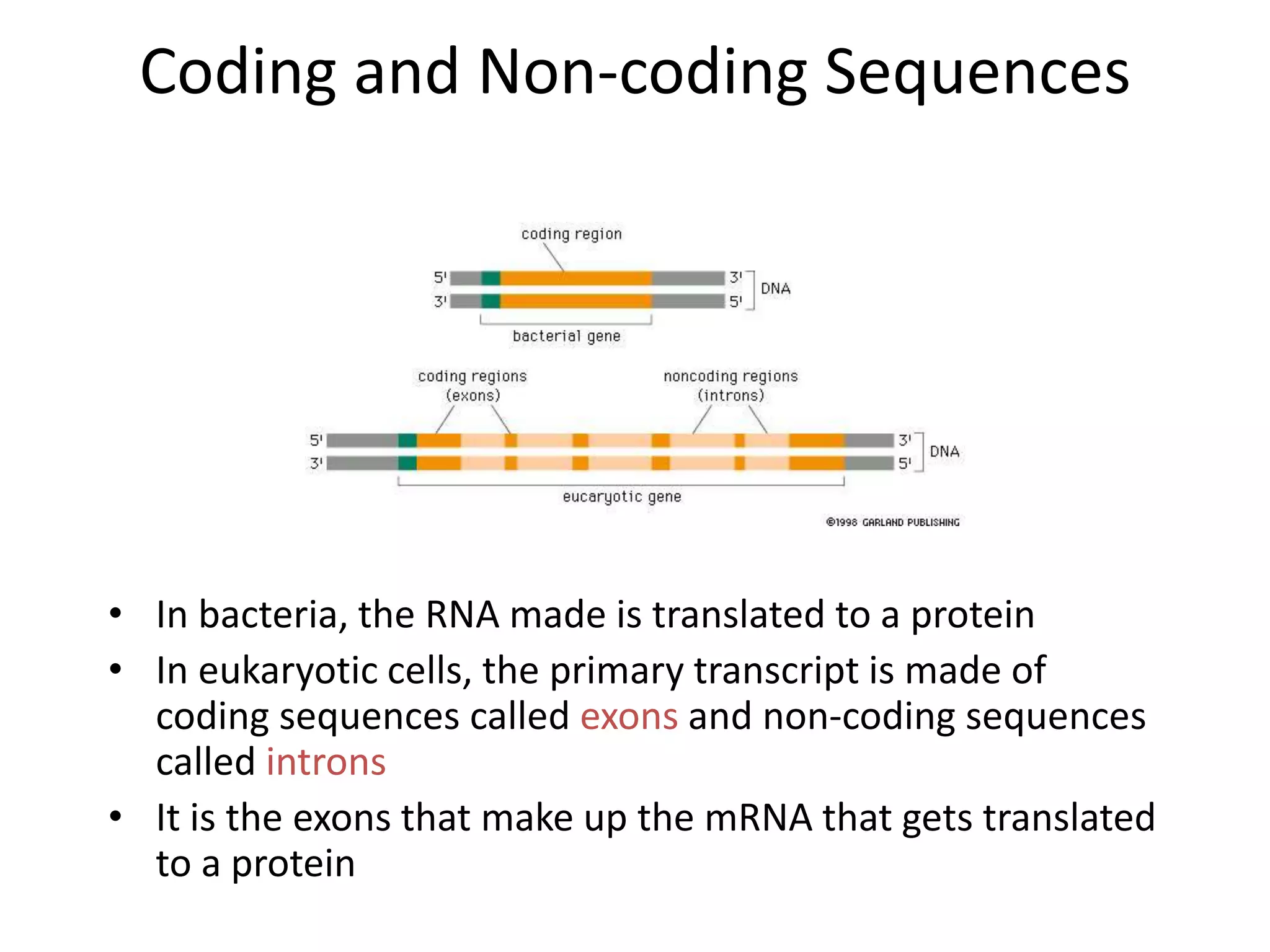
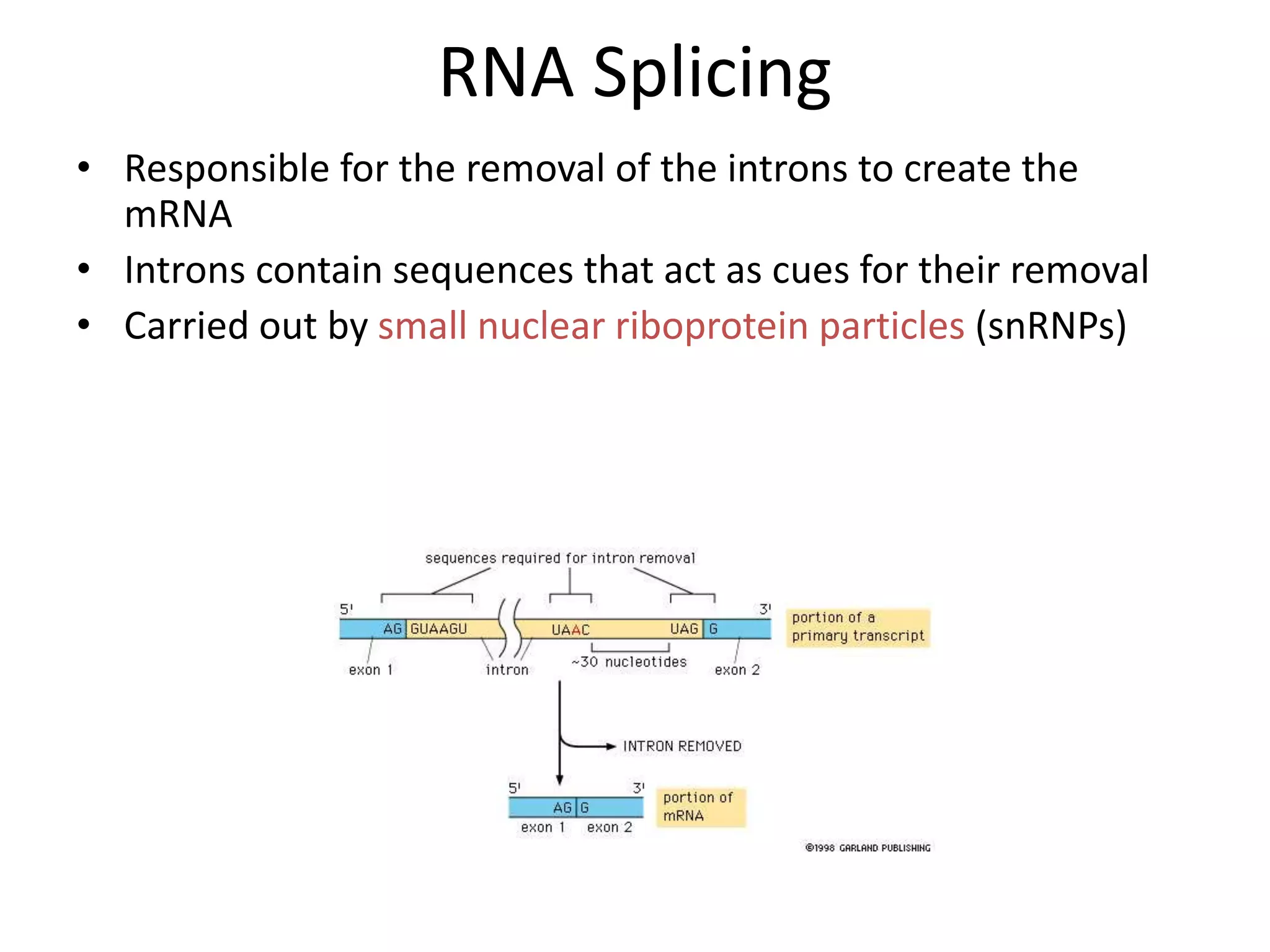
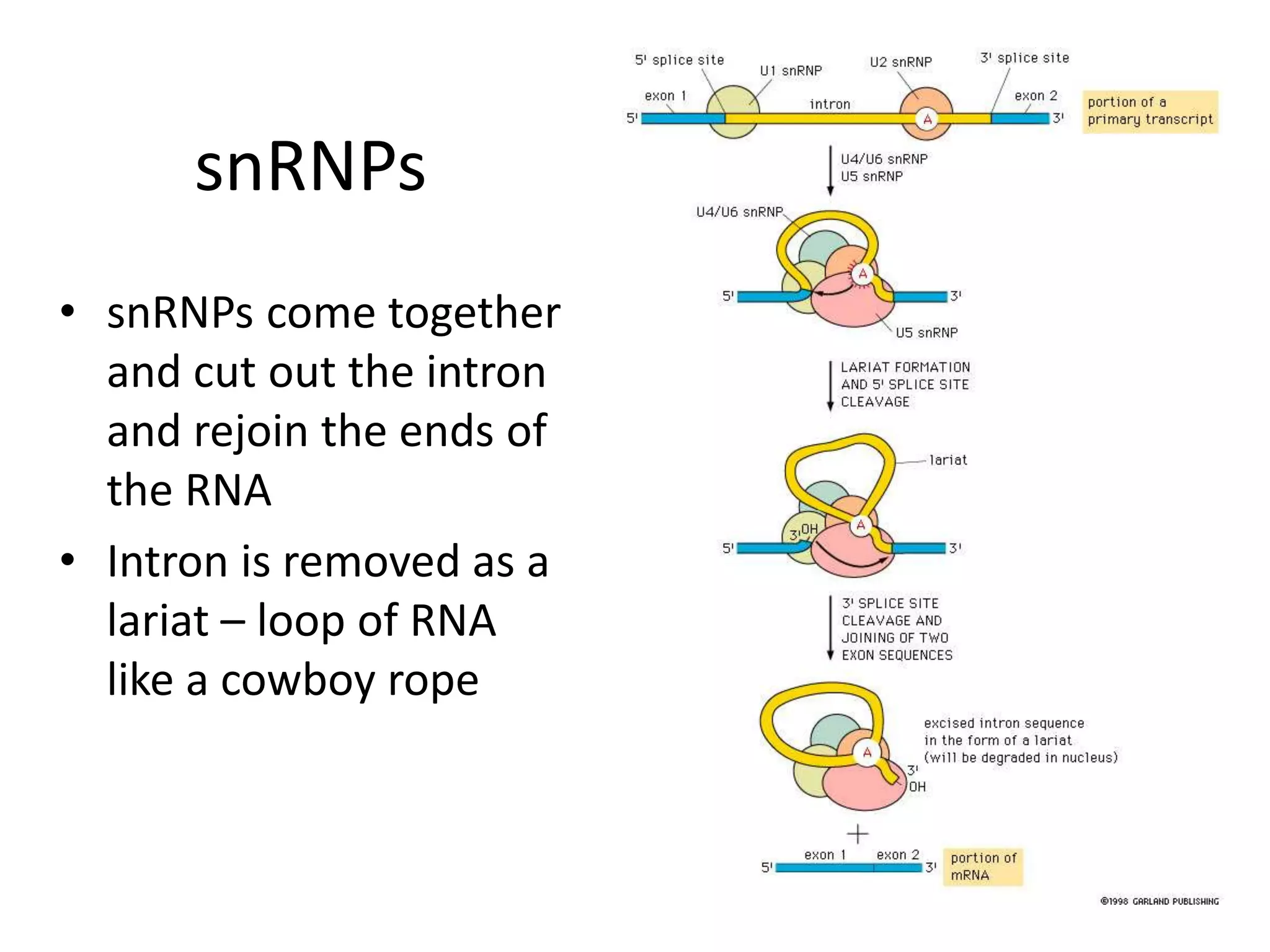
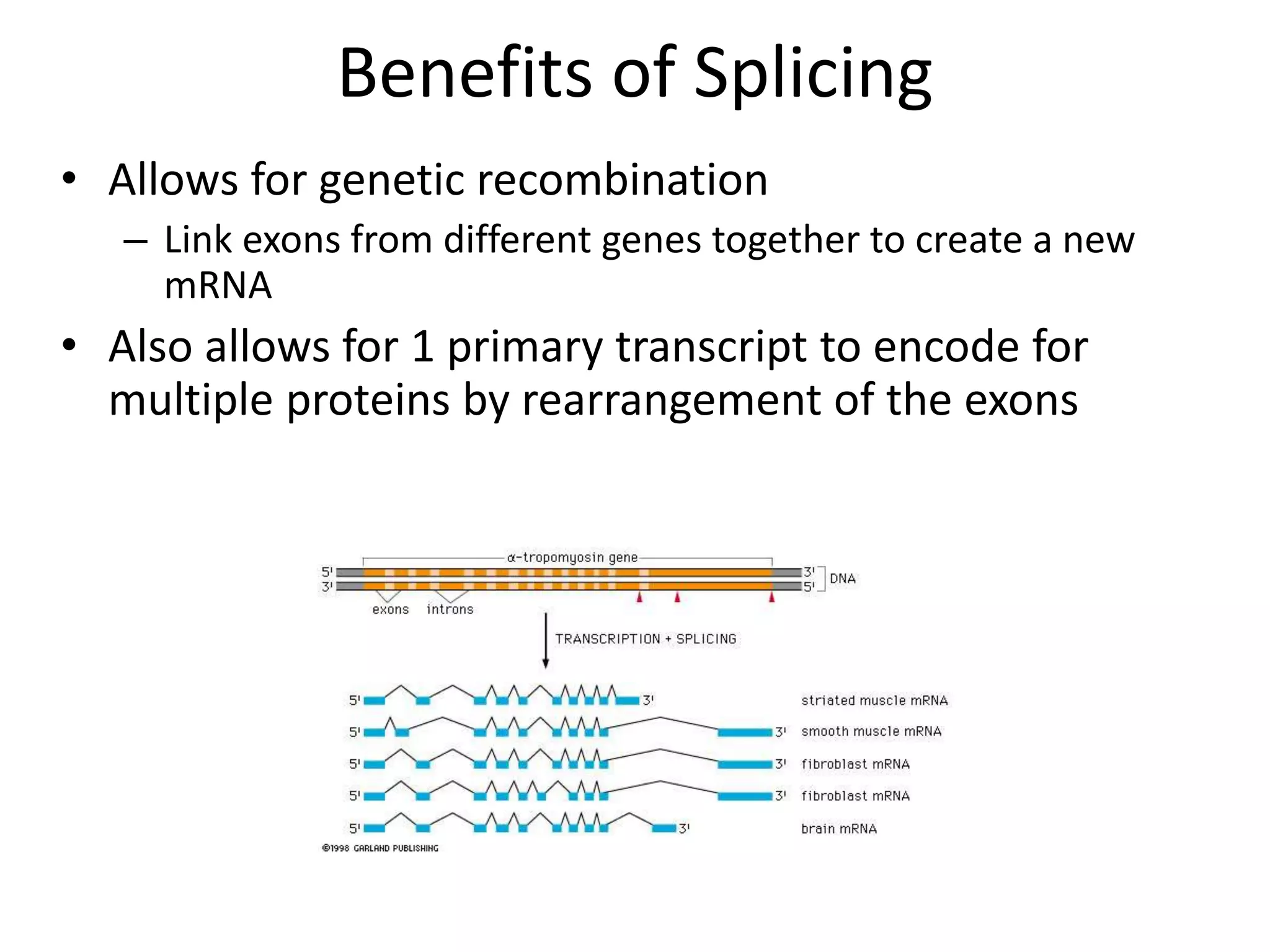
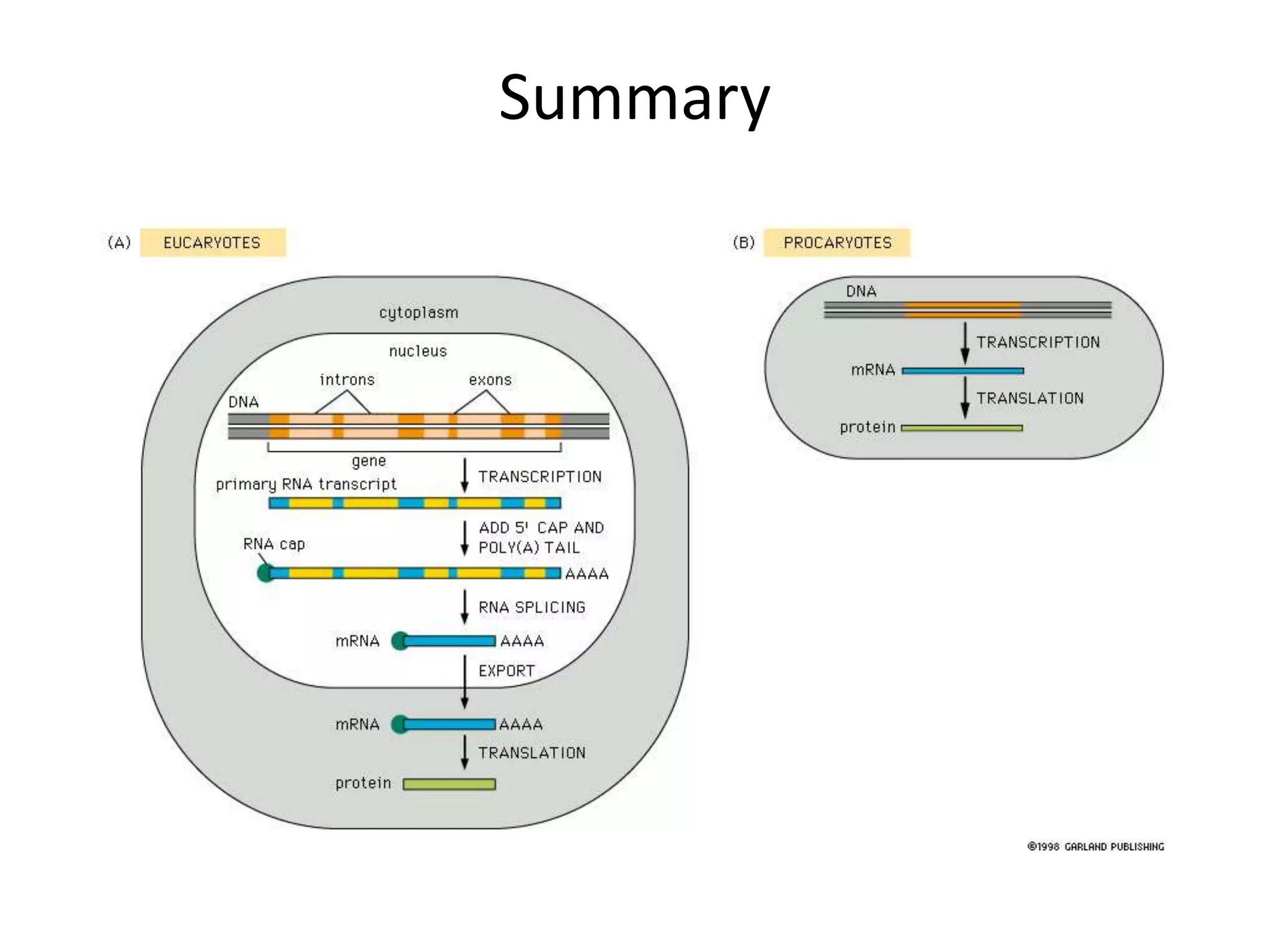
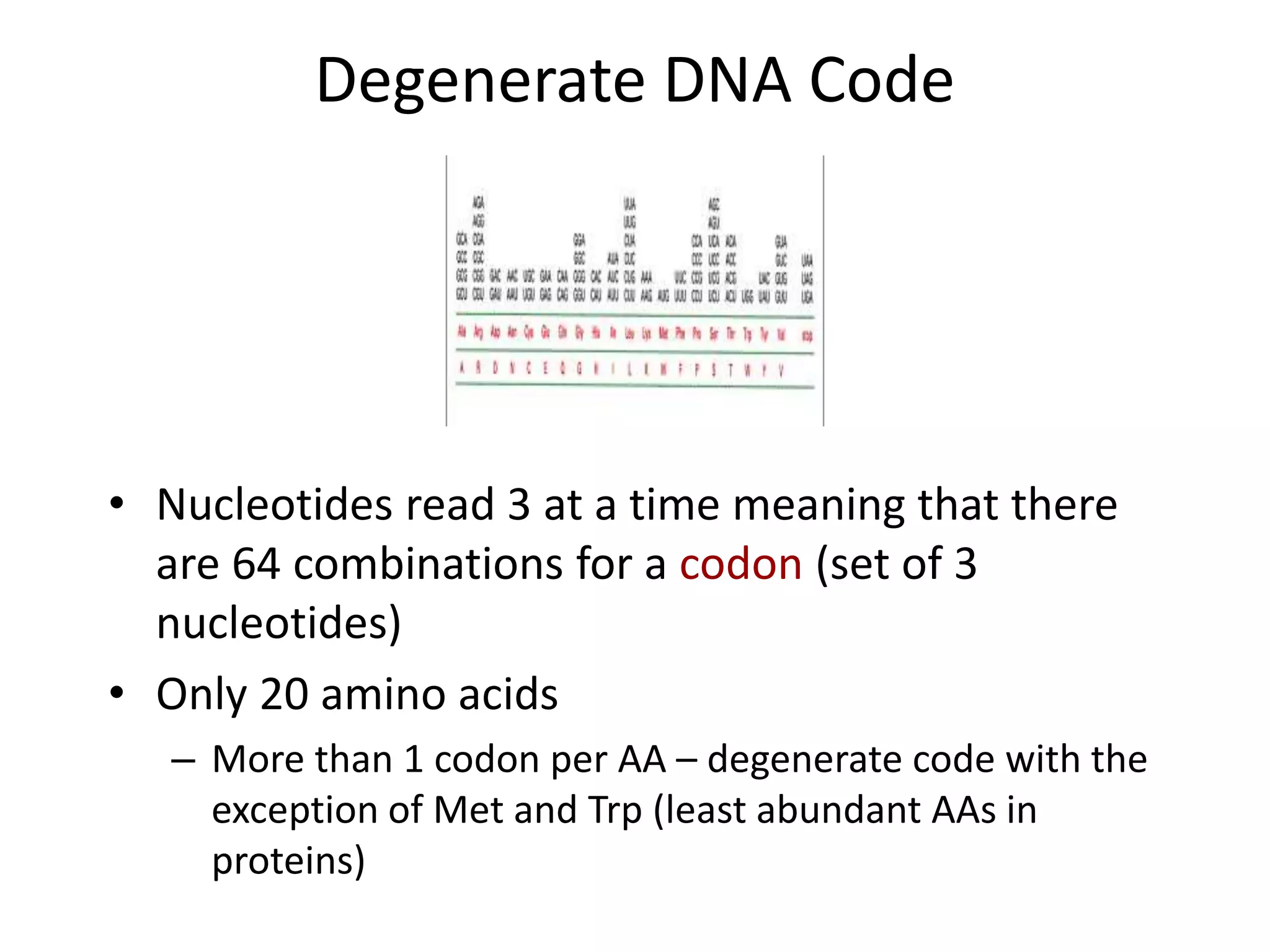
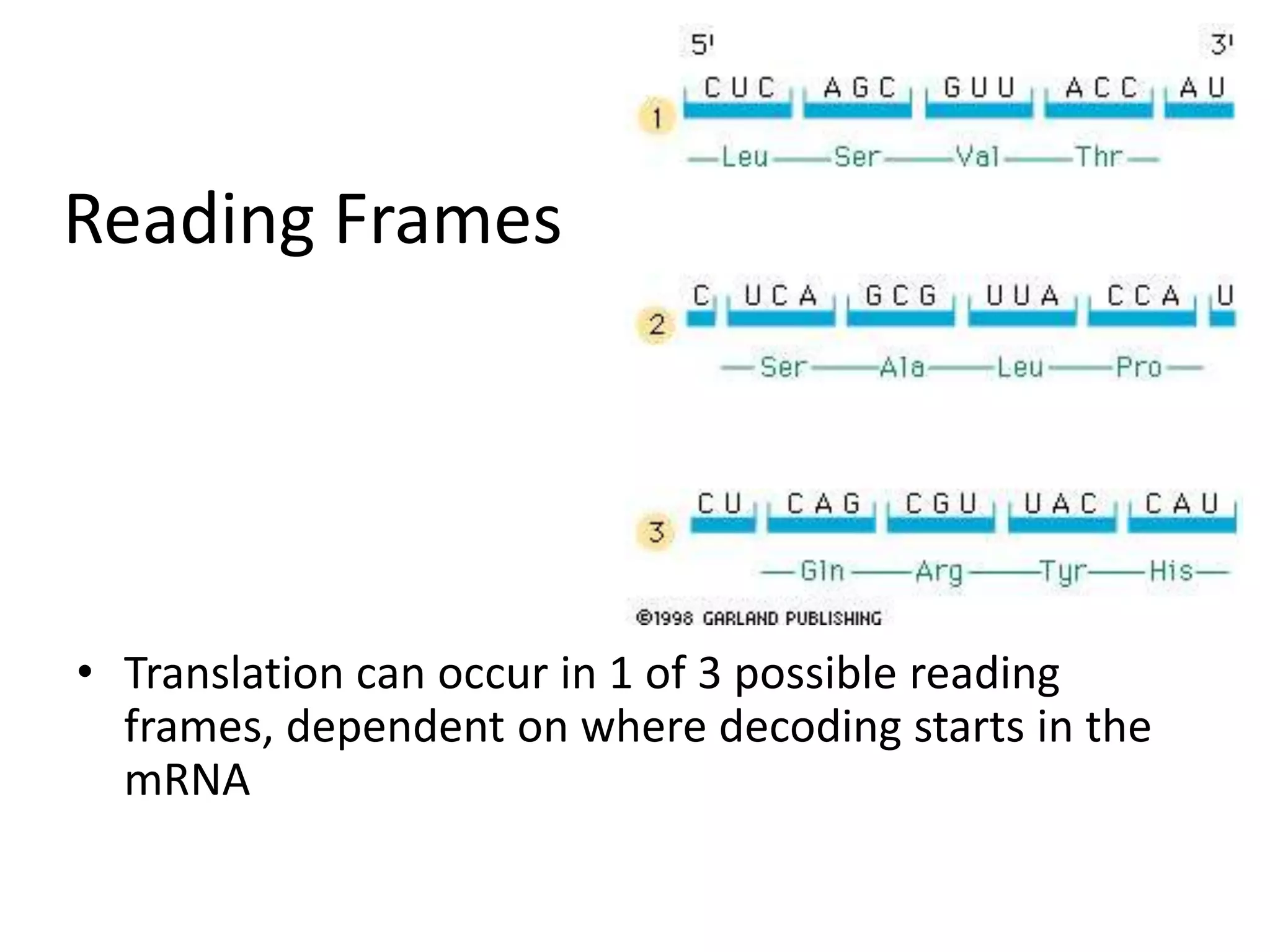
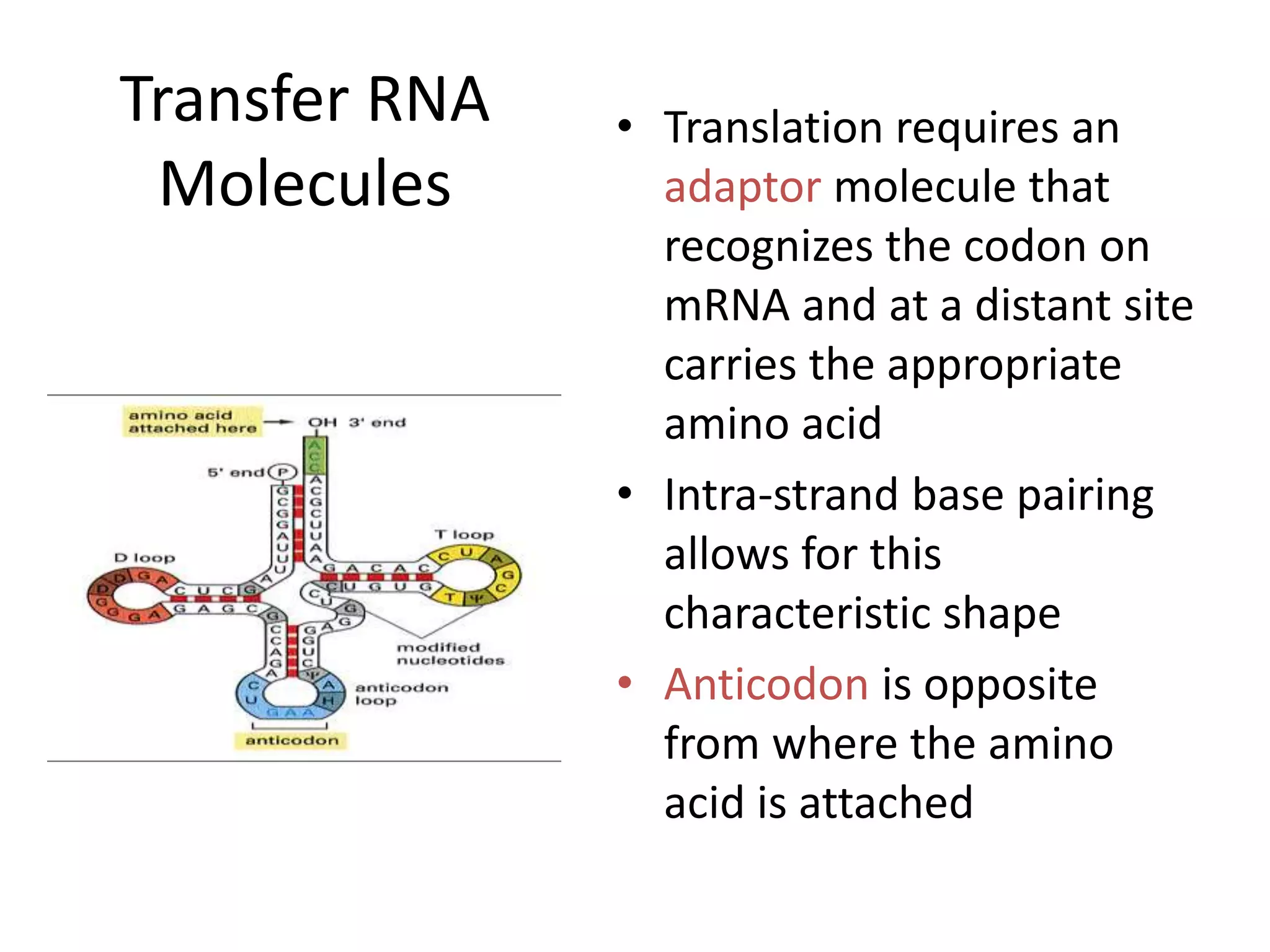
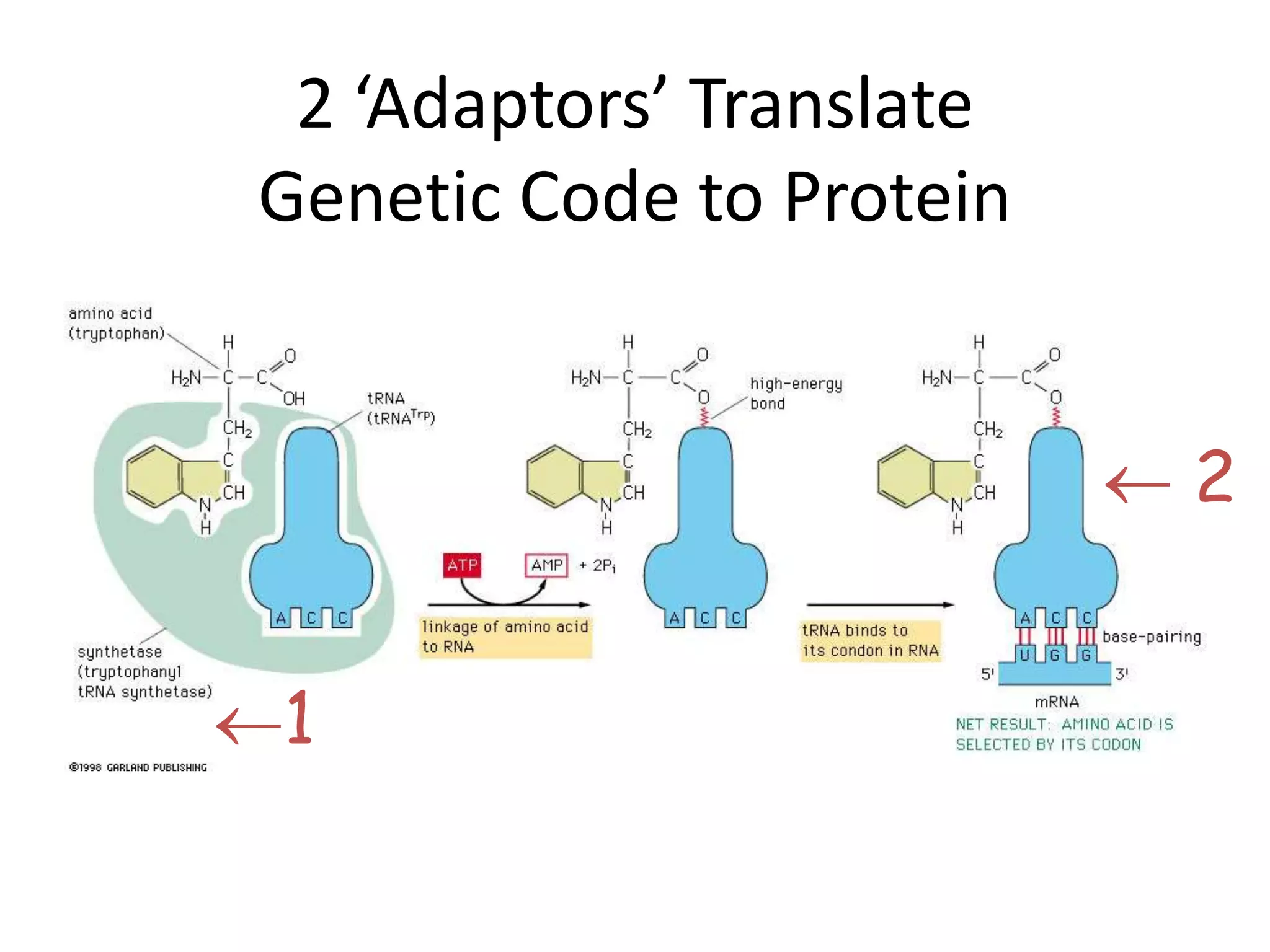
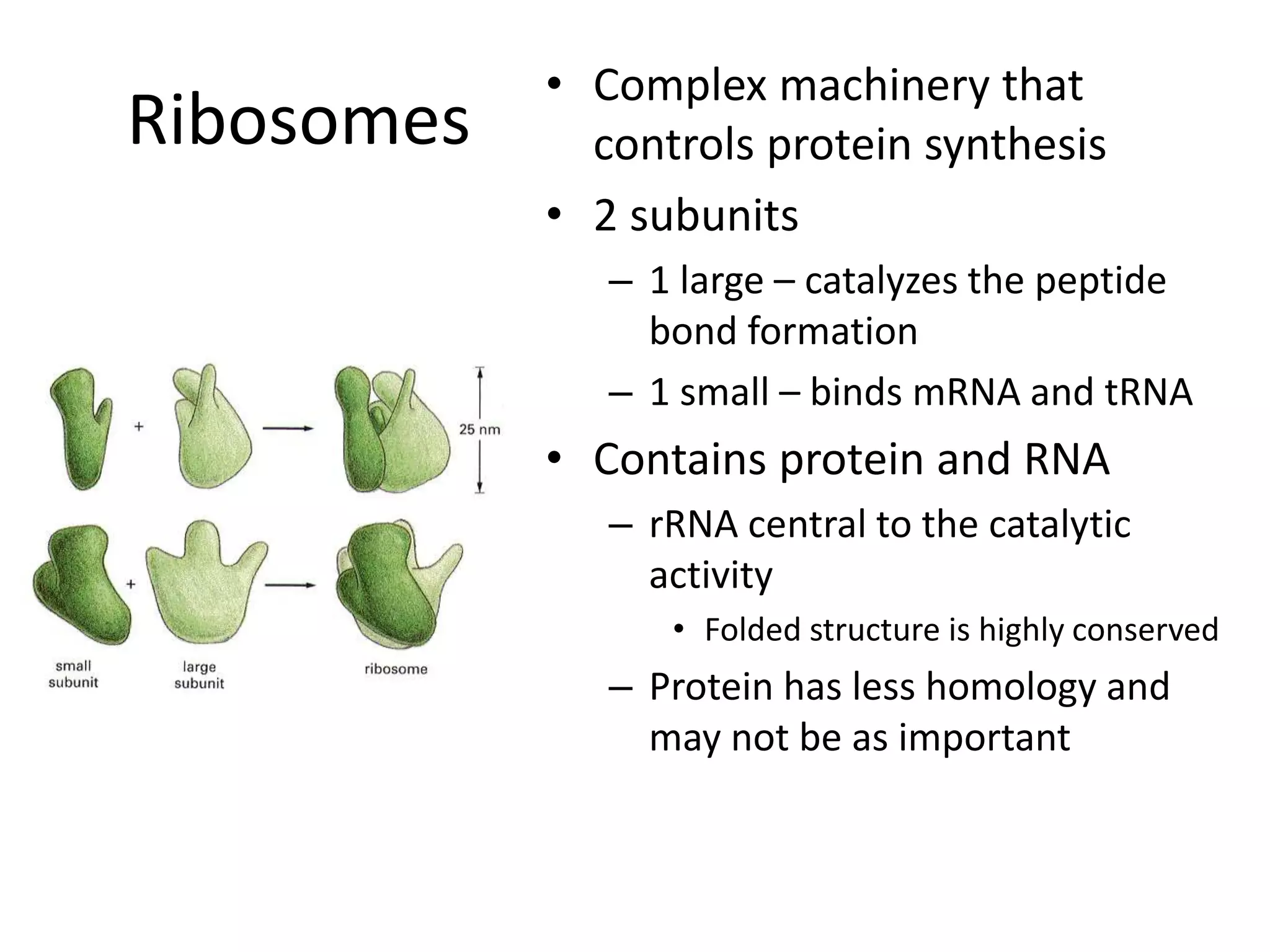
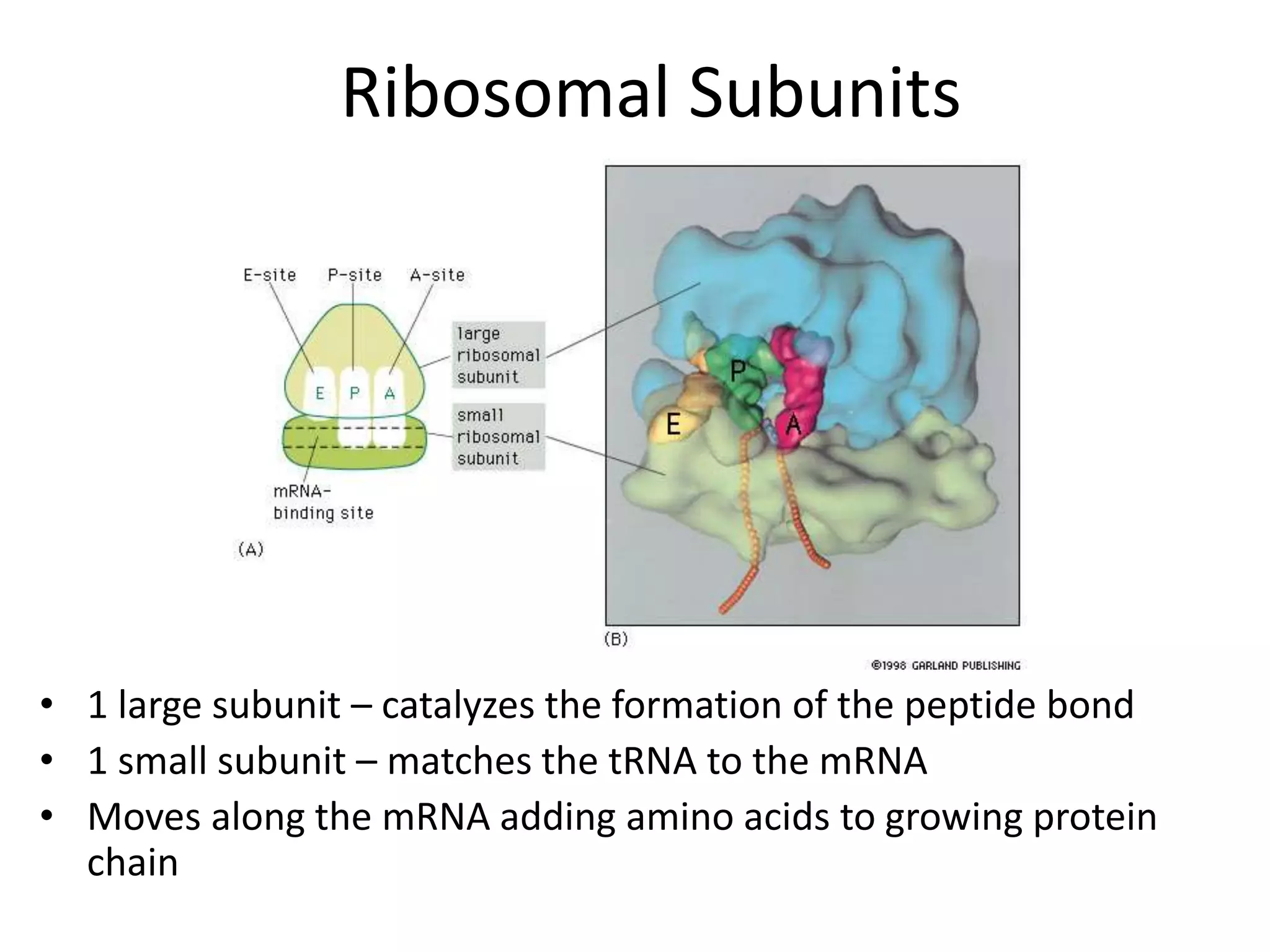
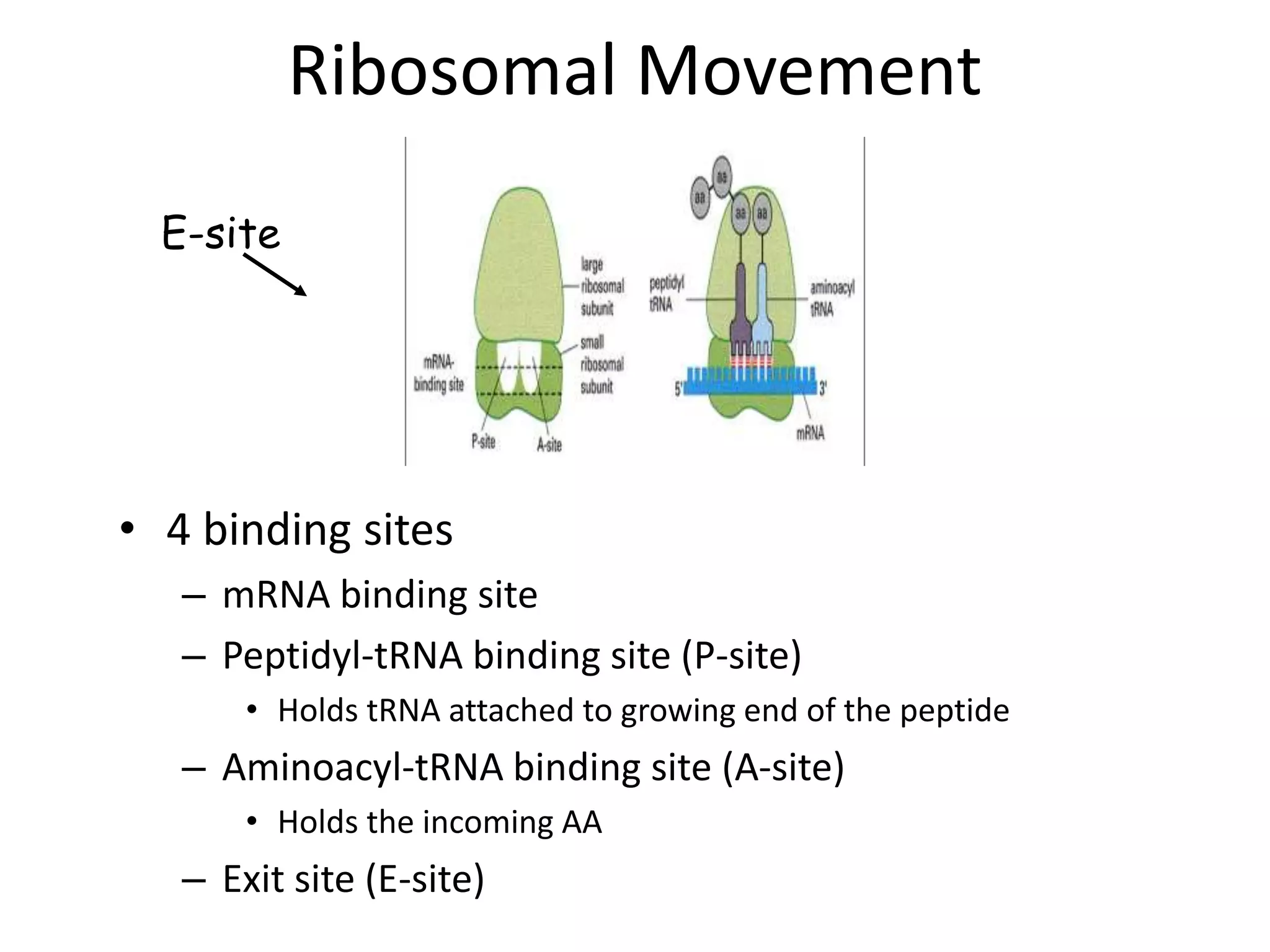
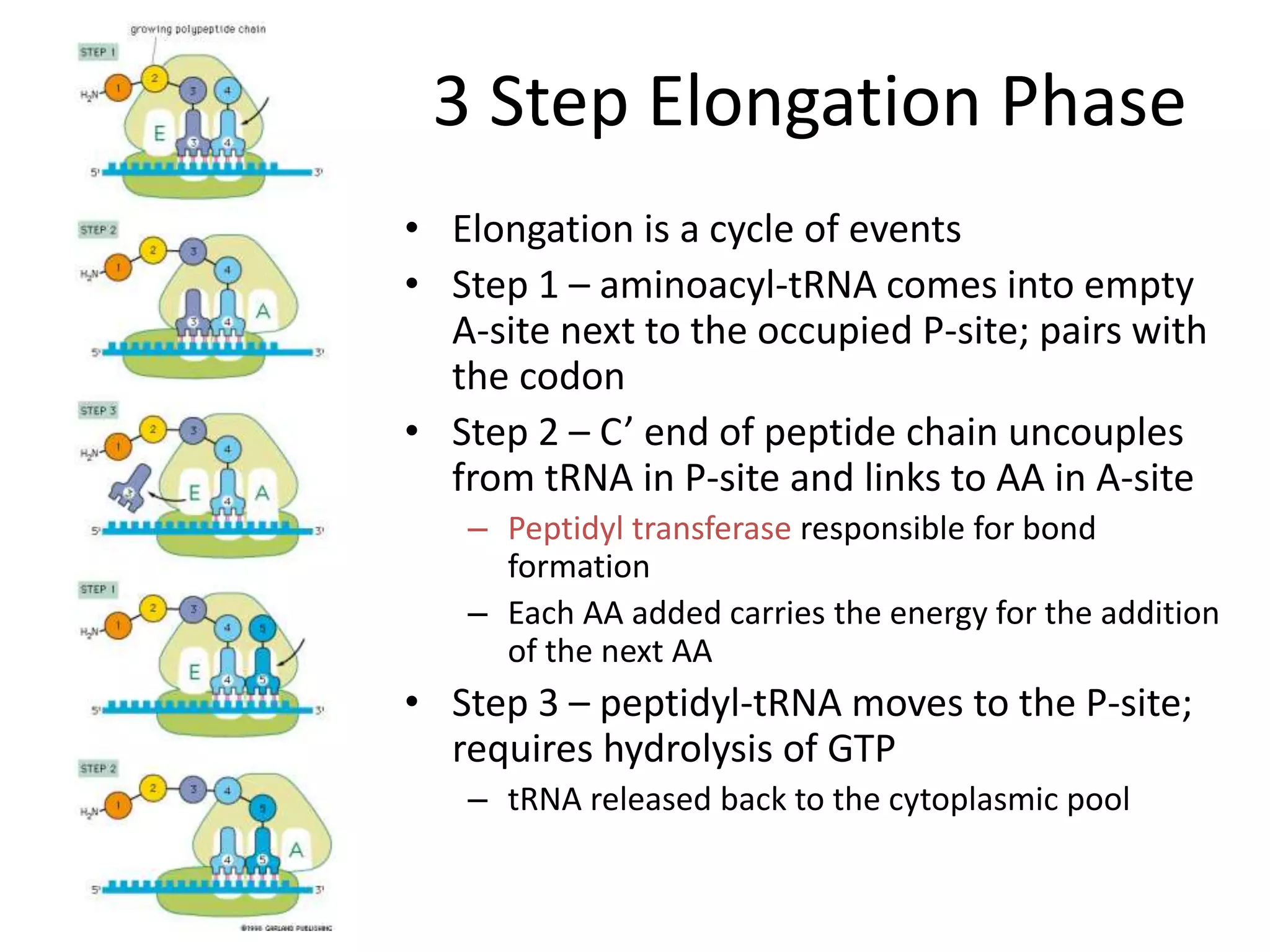
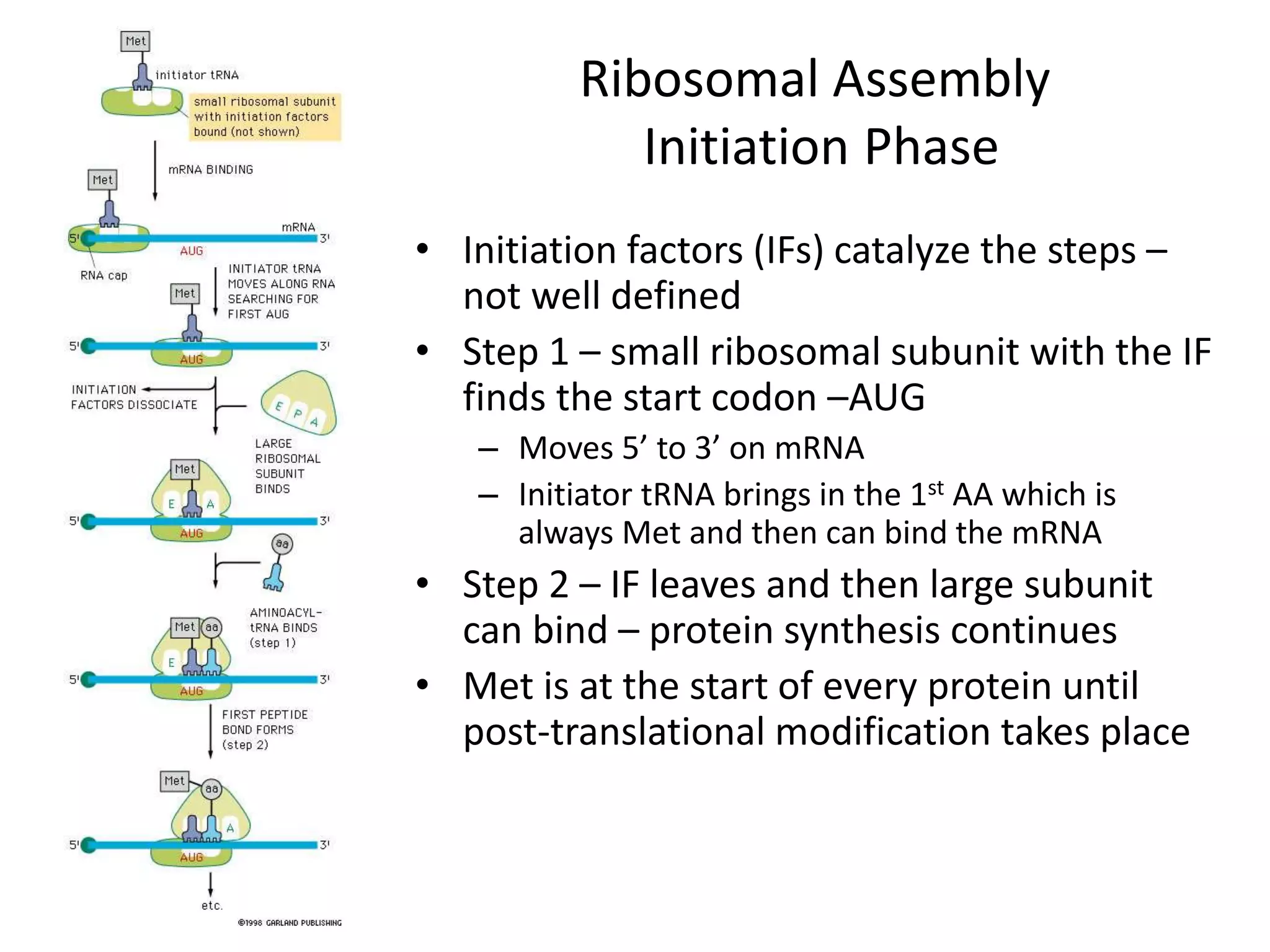
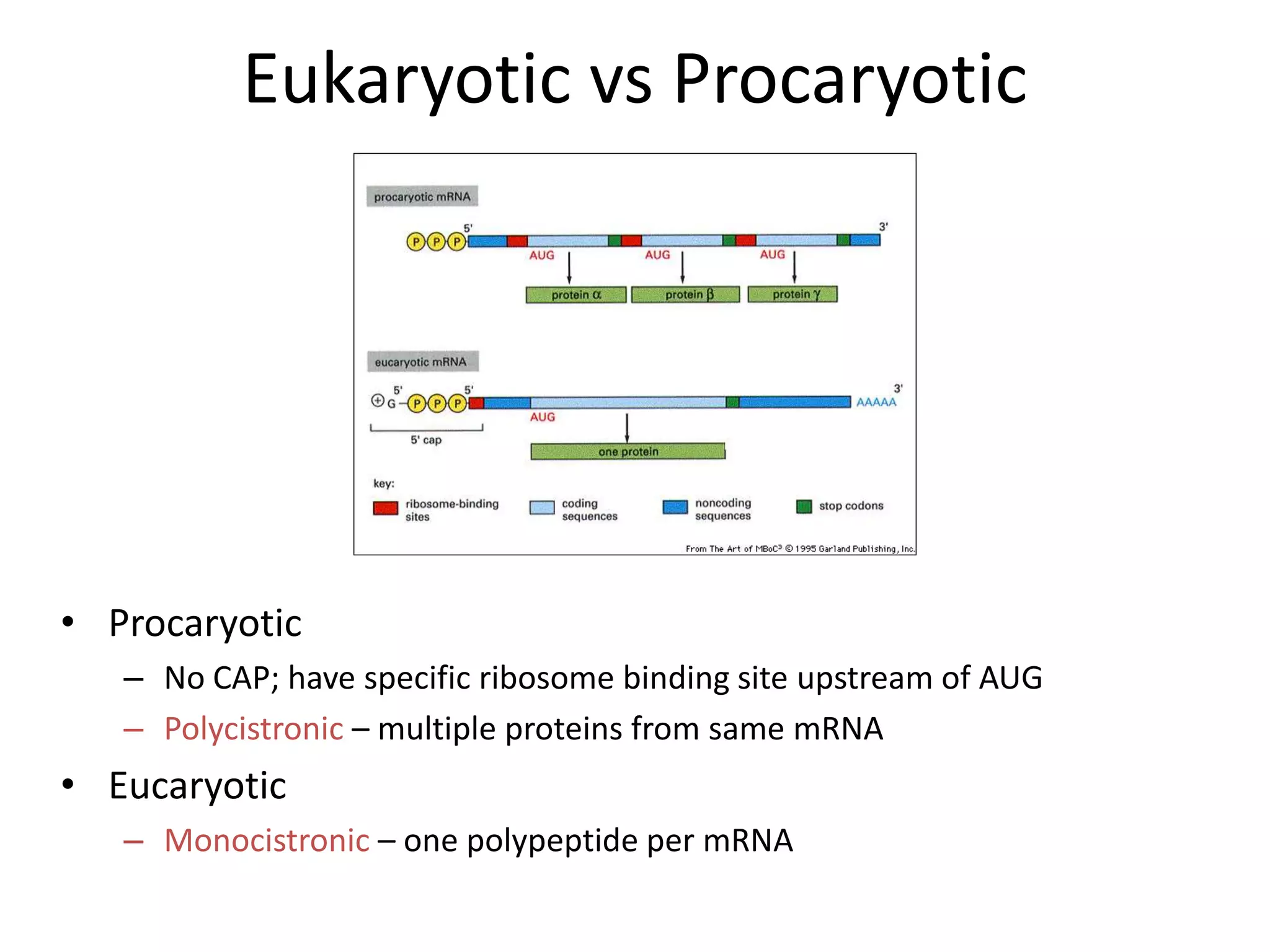
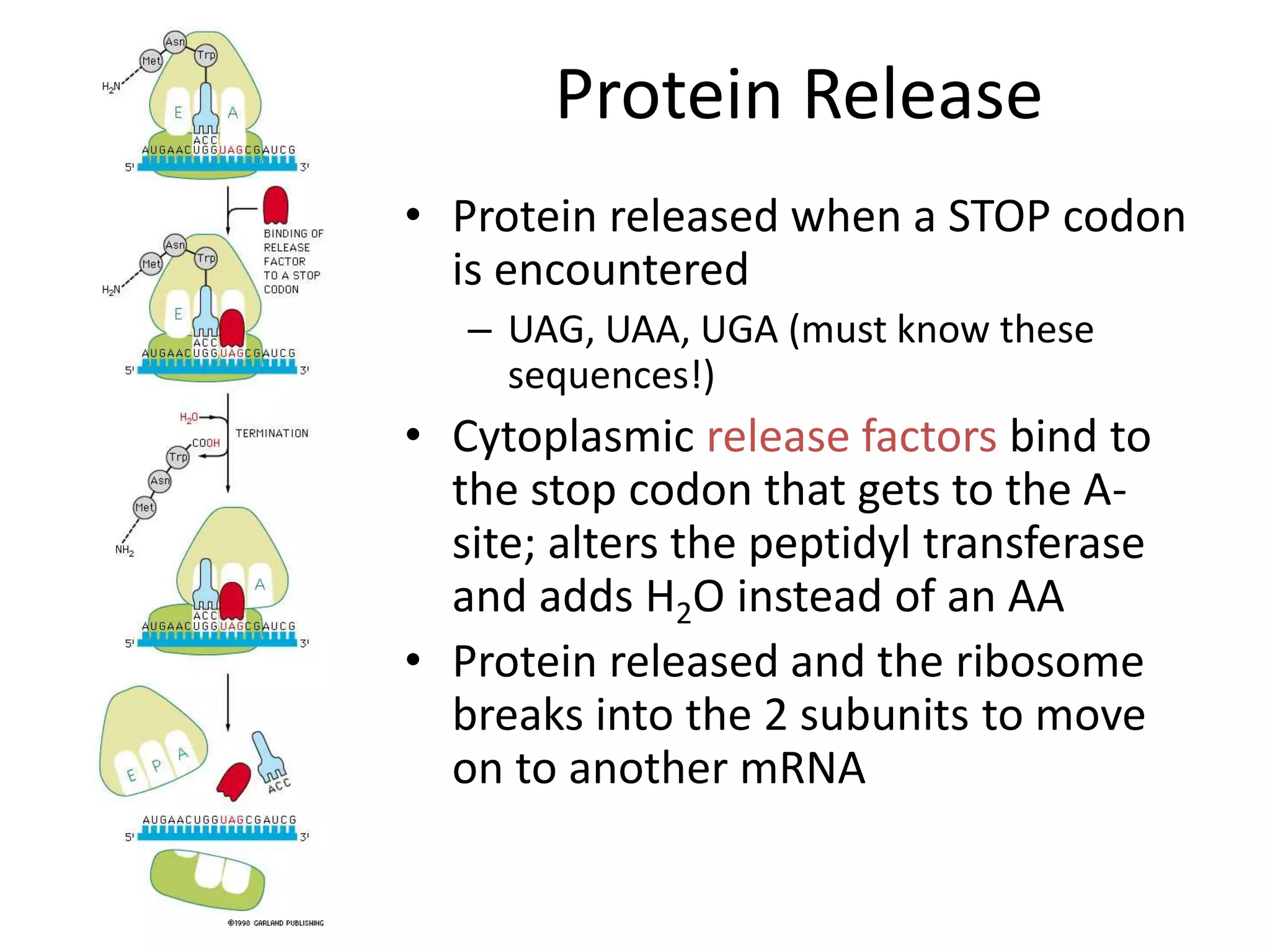
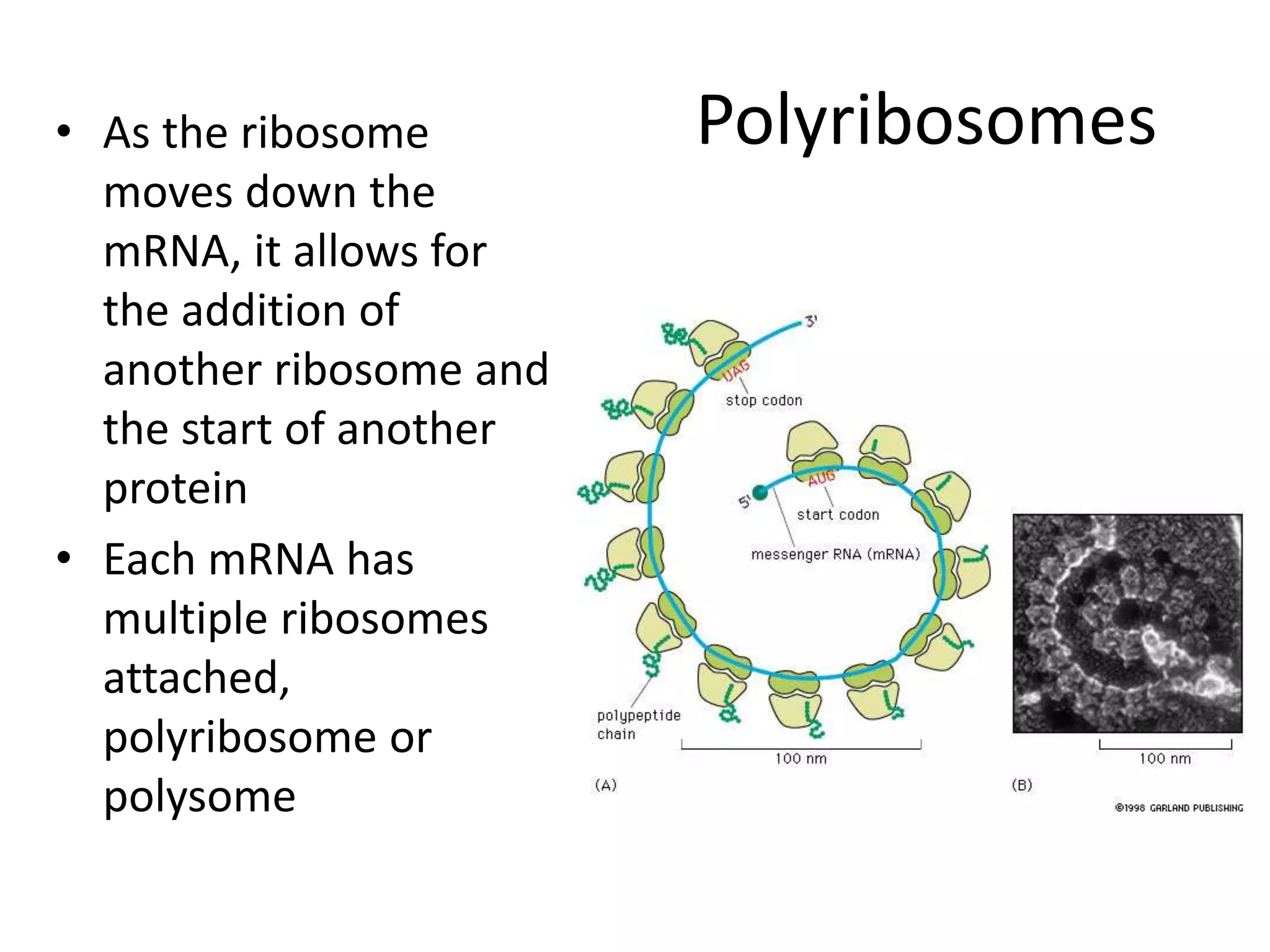
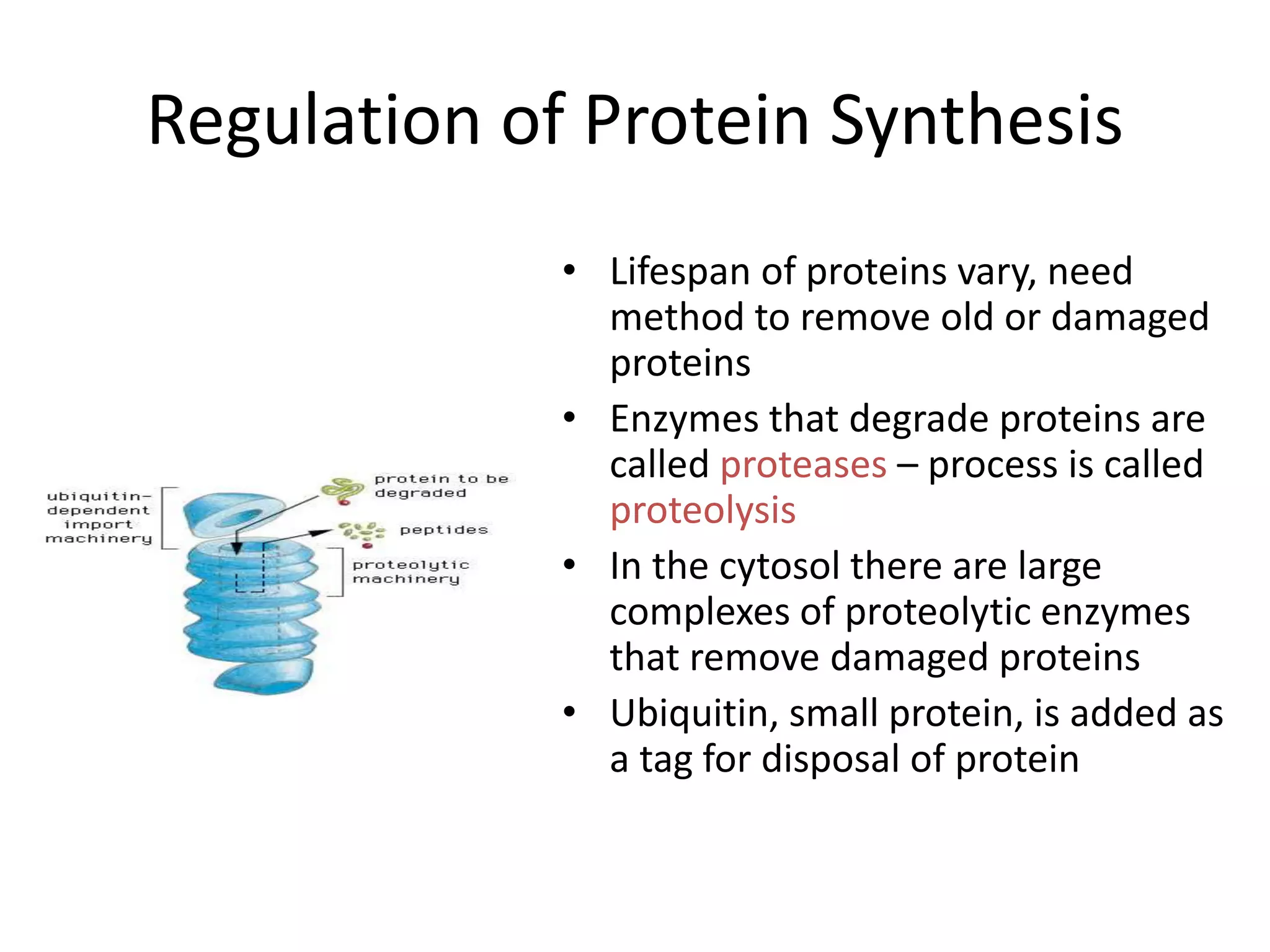
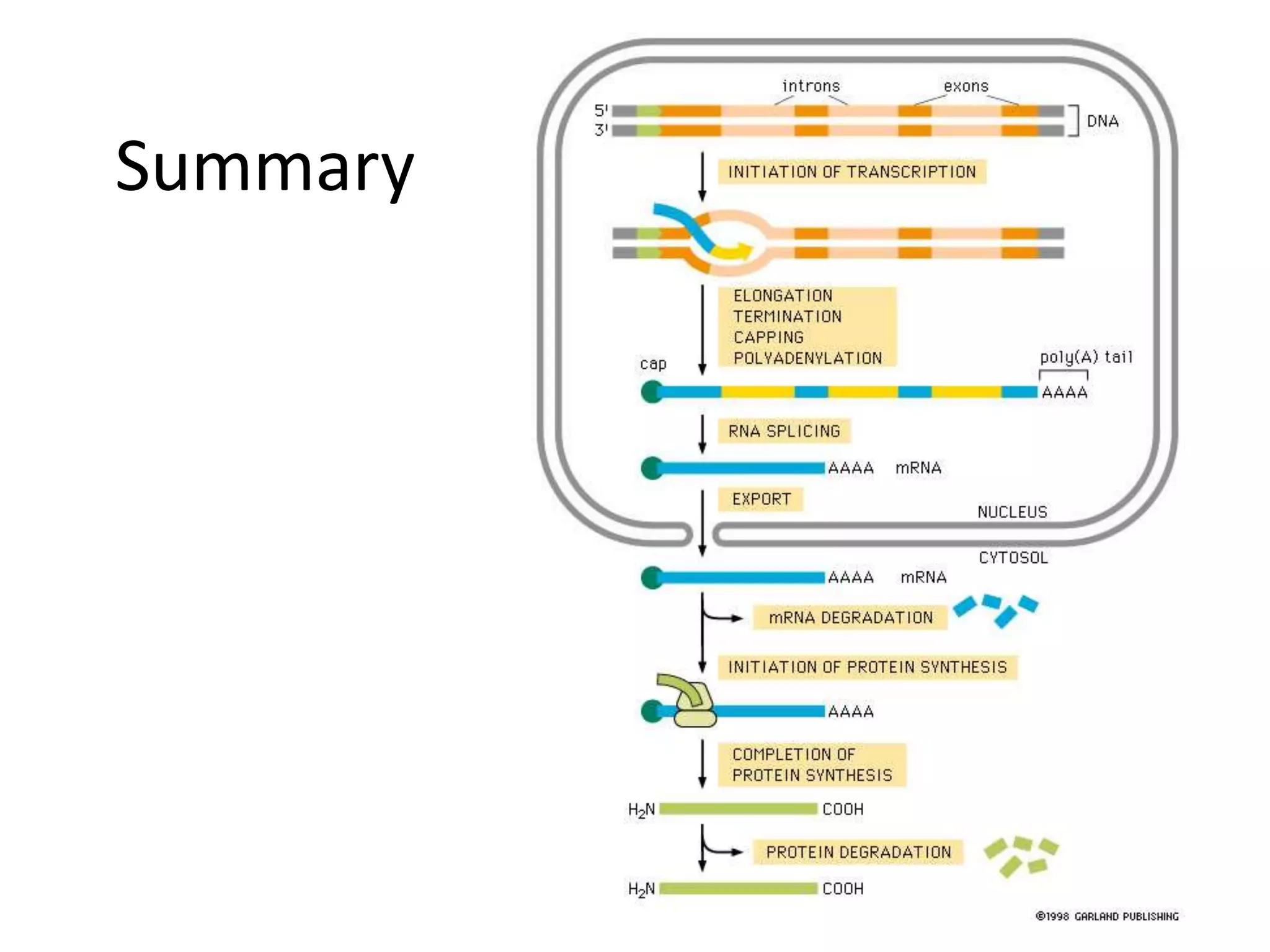
DNA acts as a template for protein production. It is transcribed into mRNA which is then translated into protein with the help of tRNA and ribosomes. Transcription occurs in the nucleus and copies DNA into RNA. Translation occurs in the cytoplasm and uses mRNA to assemble amino acids into a protein chain based on the genetic code. Gene expression can be regulated at the transcriptional or translational level to control the amount of protein produced.