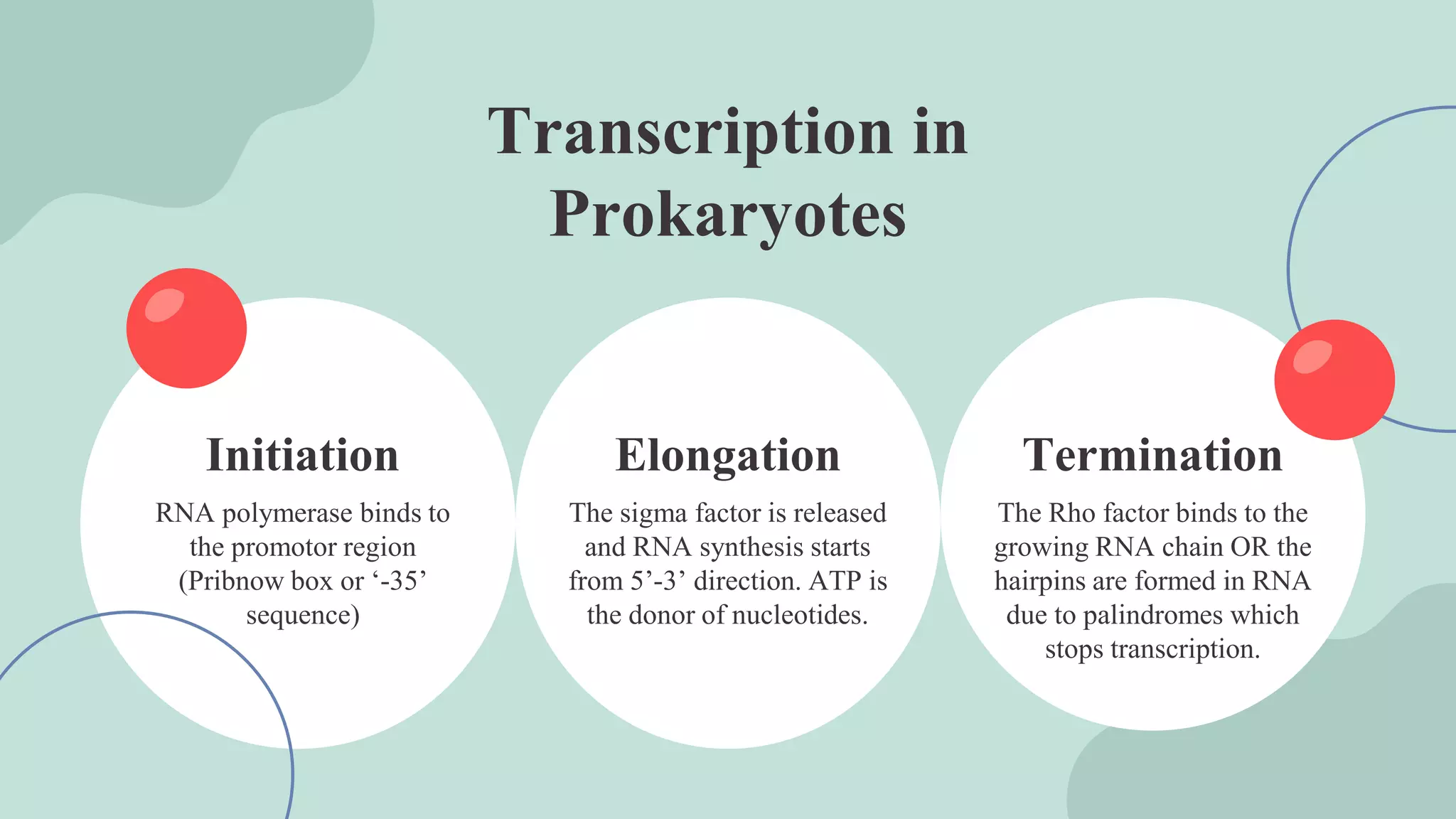
The document details the structural mechanisms of protein synthesis, focusing on DNA, RNA, and ribosomes. It explains the processes of transcription and translation, including the roles of various RNA types and ribosomal functions in producing proteins. Key points include the central dogma of molecular biology, the structure of nucleic acids, and the specifics of ribosome assembly and function.