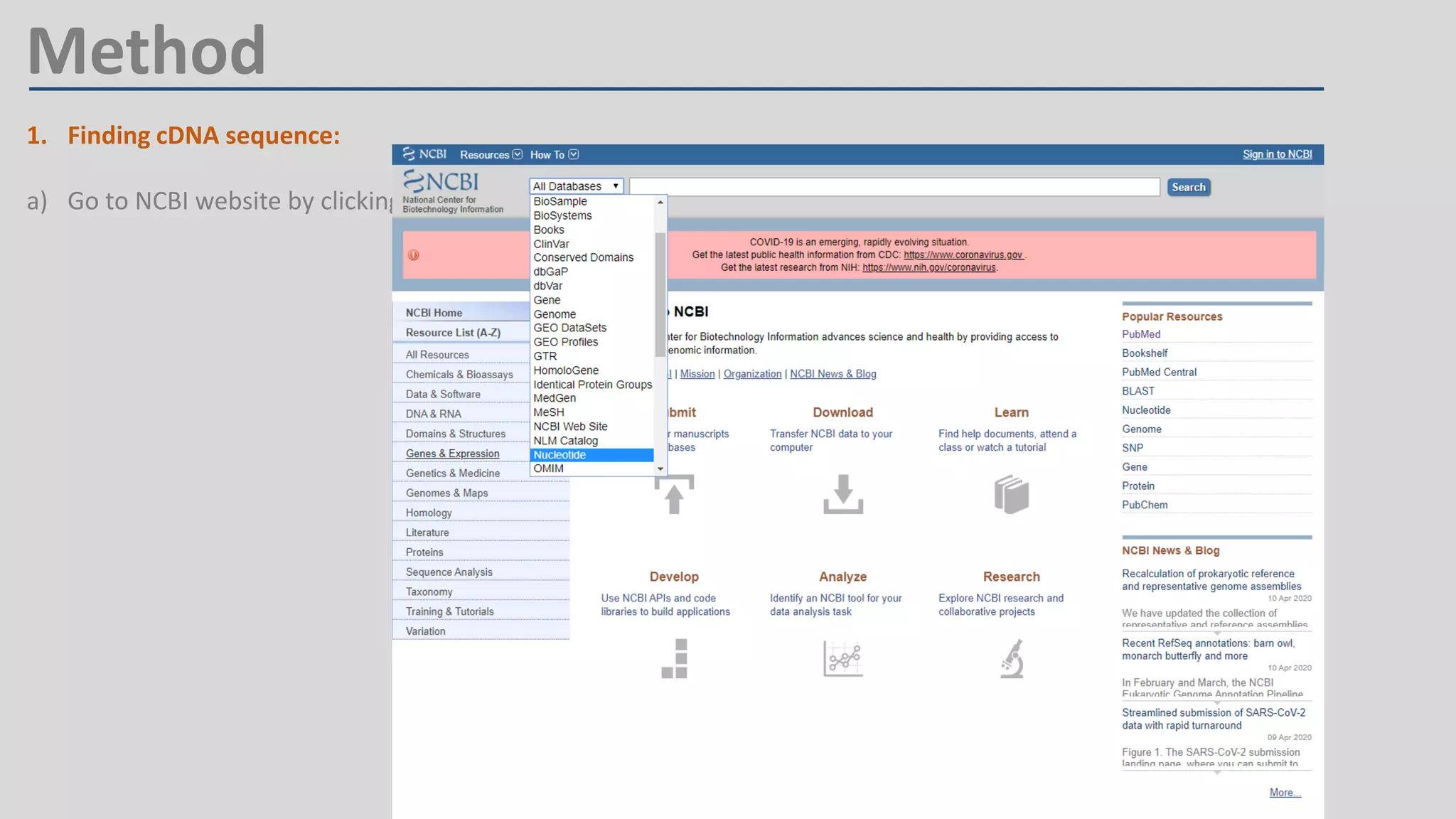
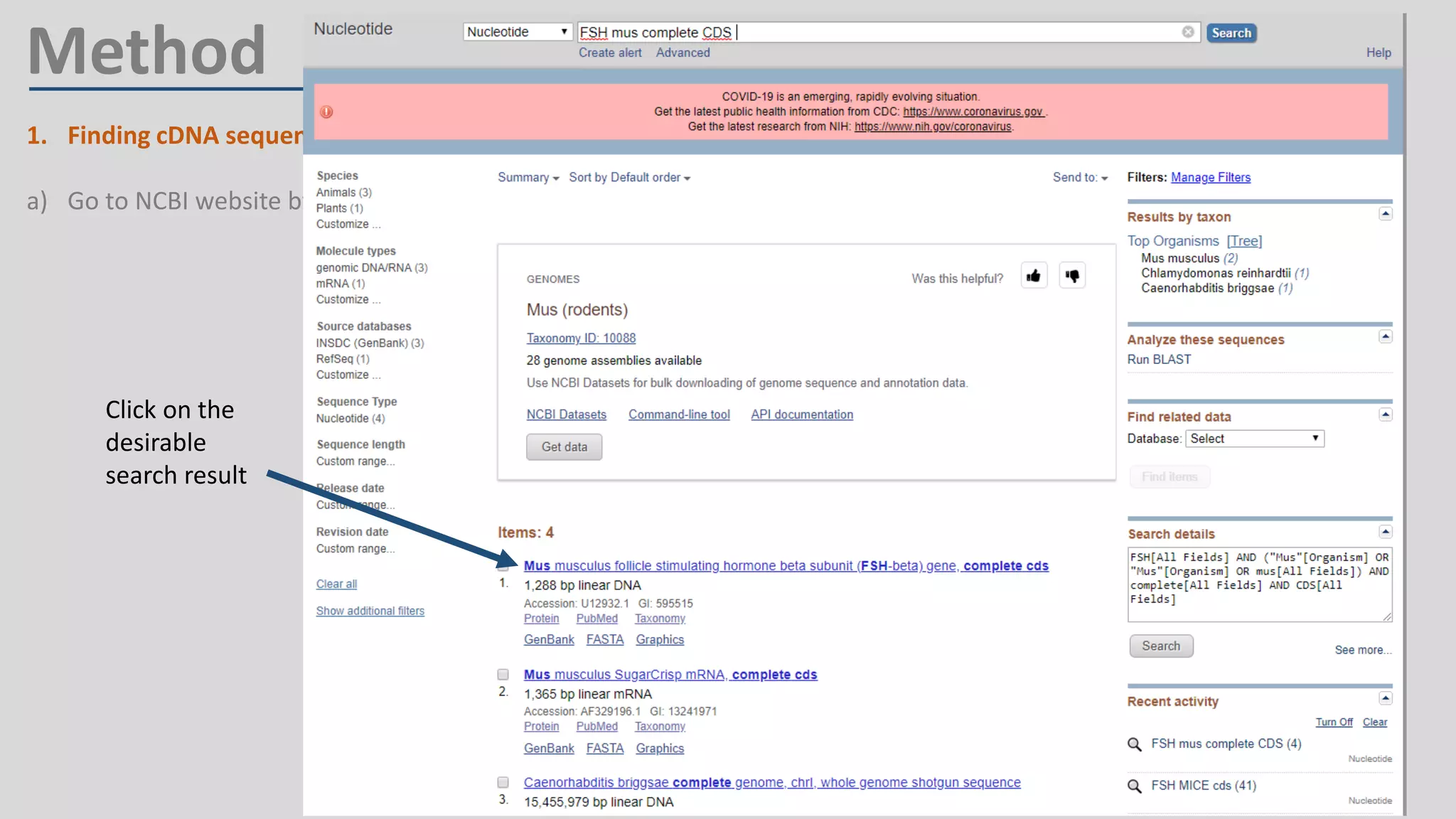
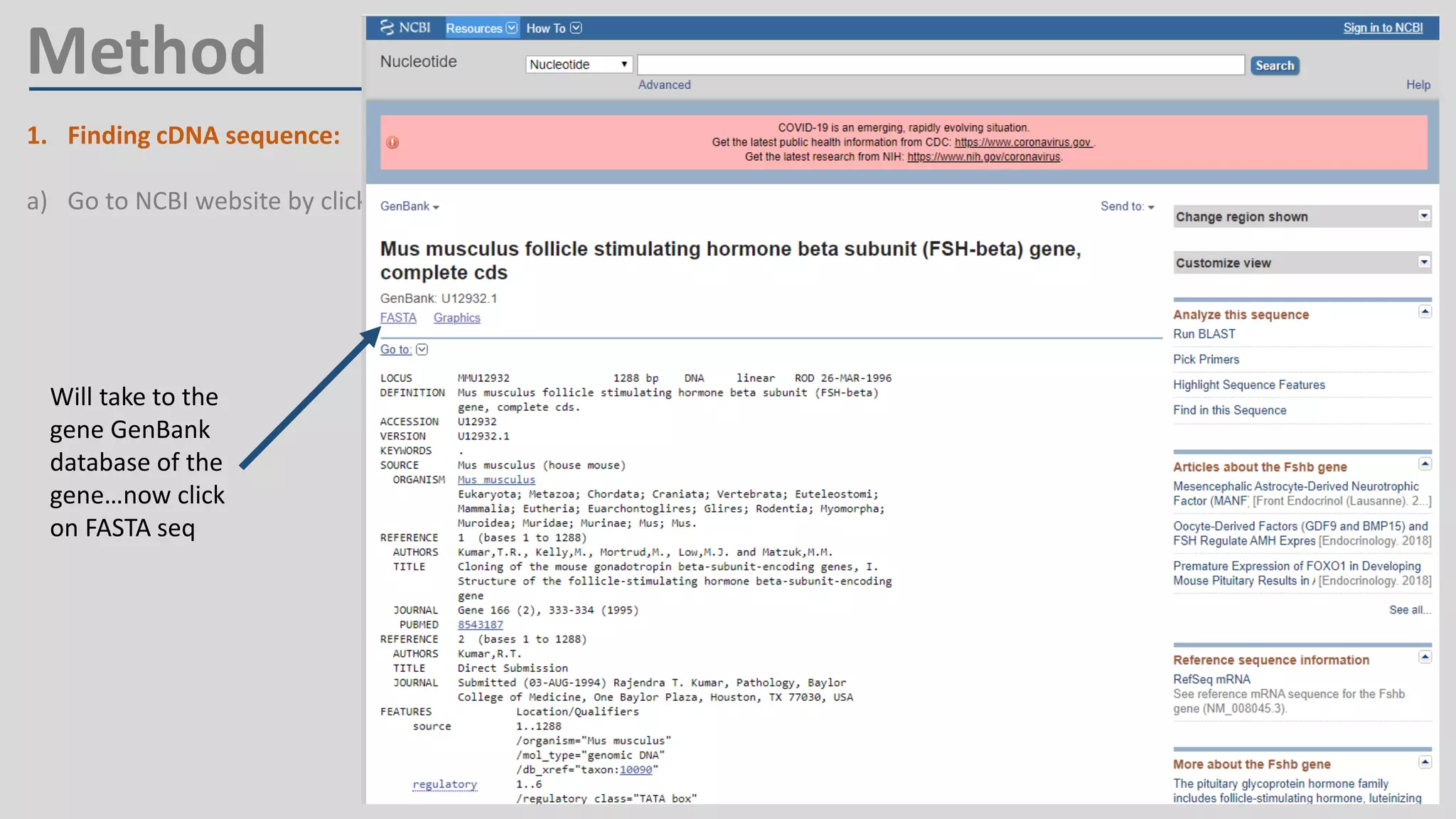
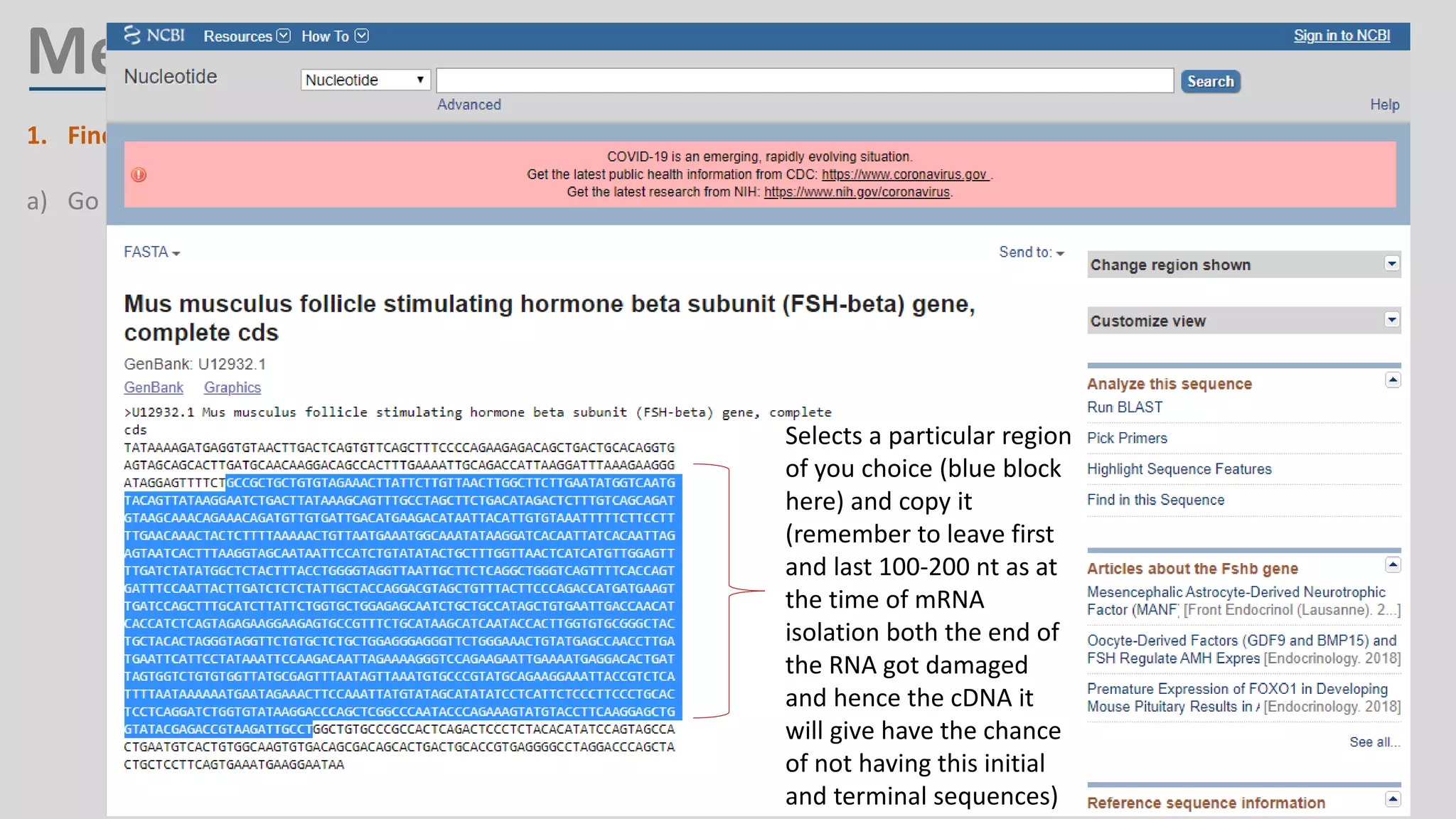
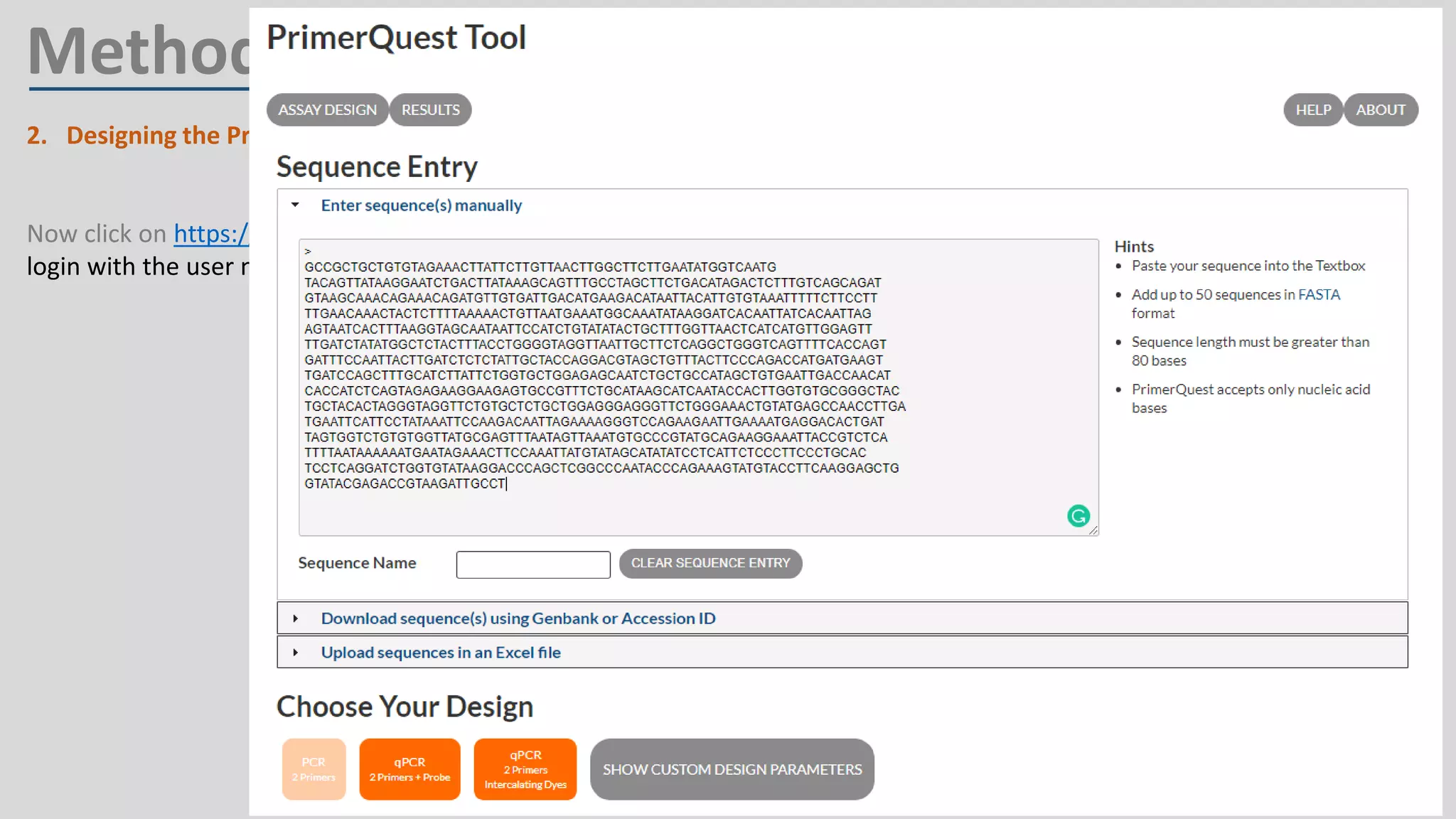
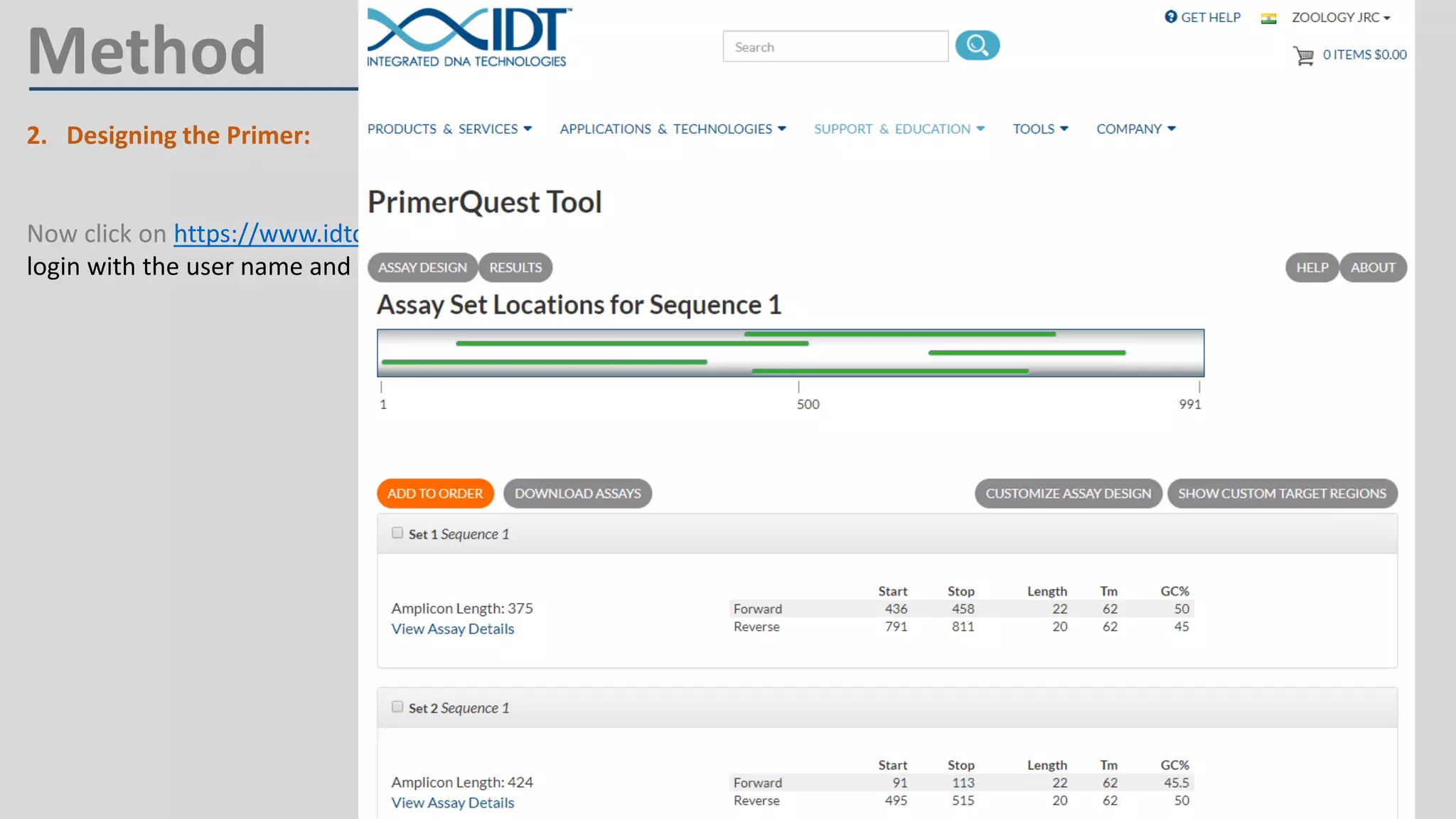
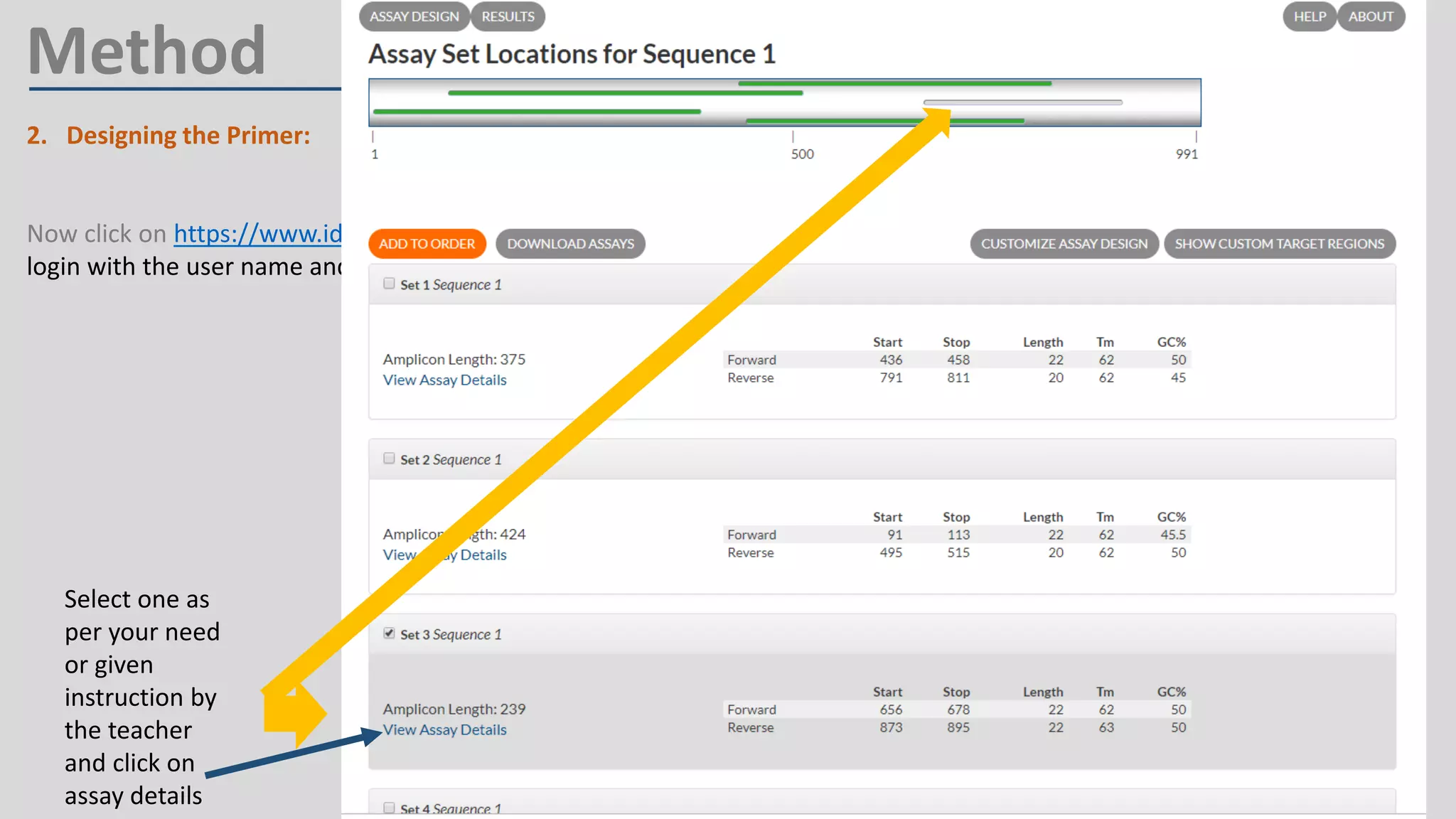
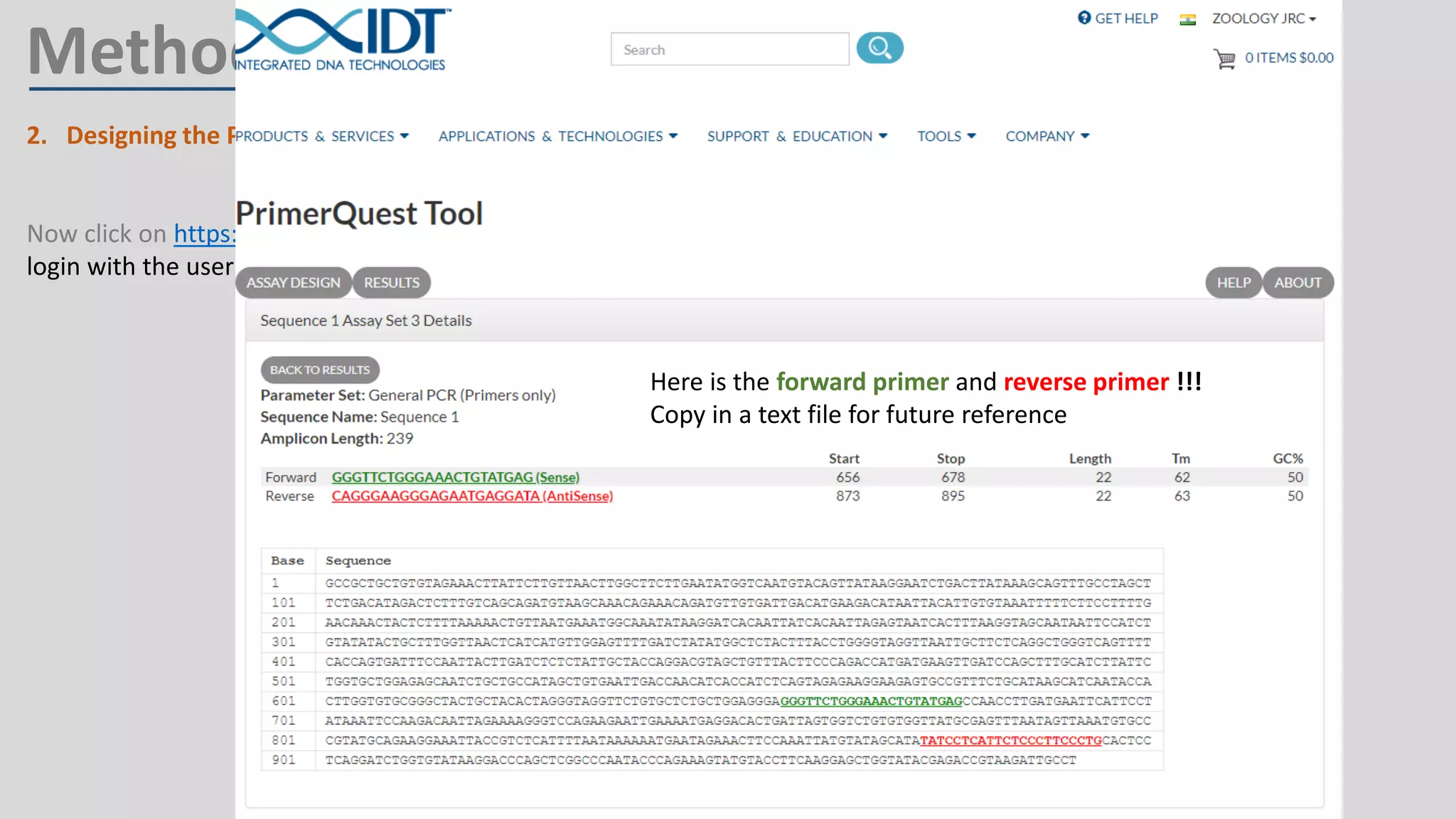
Primer design involves finding the cDNA sequence of interest from NCBI, pasting it into the PrimerQuest tool on IDT to generate primer pairs that meet criteria like length 18-25 bp, GC content 40-60%, and no complementarity or repeats exceeding 3 bp; the best primer pair is then selected and their sequences copied for future PCR experiments.