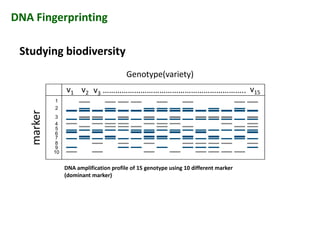
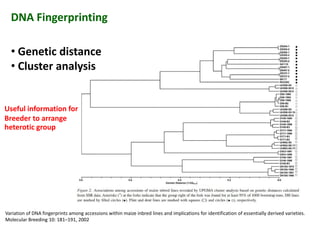
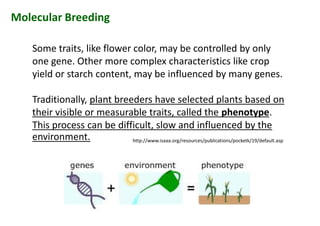
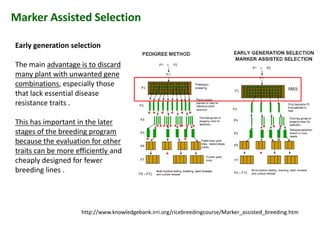
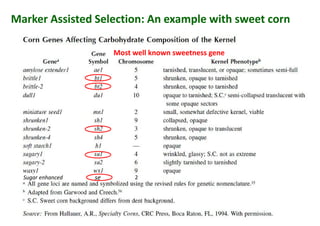
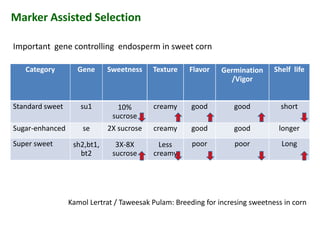
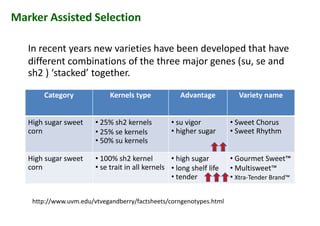
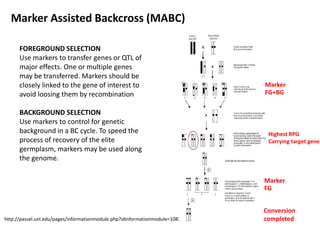
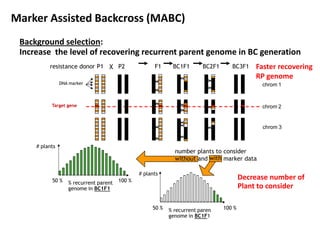
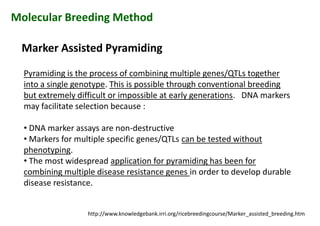
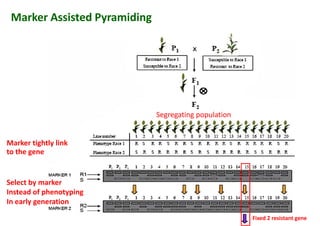
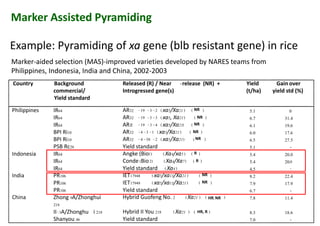
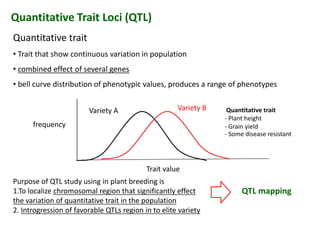
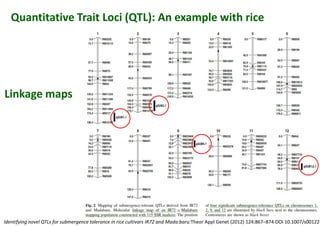
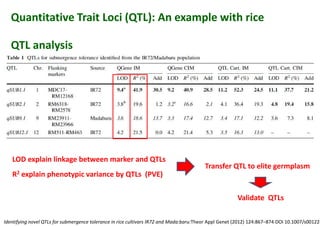
The document discusses molecular breeding and marker-assisted selection (MAS) in crops, outlining various techniques such as DNA fingerprinting, marker-assisted backcrossing (MABC), and genomic selection. It highlights the advantages of using molecular markers over traditional phenotype selection, including early selection and reduced costs, to improve desirable traits in plant breeding. Additionally, the document explores the processes of pyramiding multiple genes to enhance characteristics like disease resistance and crop yield.