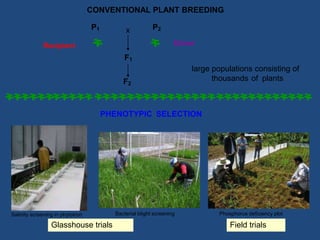
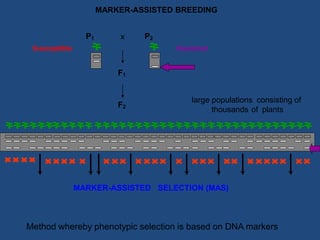
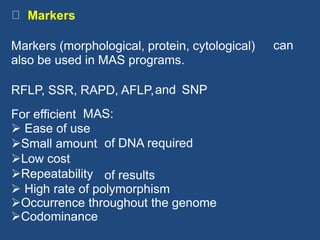
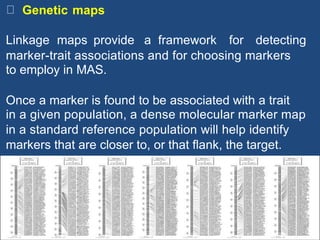
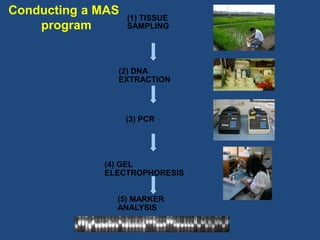
Marker-assisted selection (MAS) is a plant breeding method that uses molecular markers linked to traits of interest to guide selection for desirable crop traits. It allows breeders to select plants earlier in the breeding cycle compared to phenotypic selection alone. MAS has advantages like being unaffected by environmental conditions and enabling selection for recessive traits. However, it also has drawbacks like being more expensive than conventional techniques and the possibility of recombination between markers and traits. Careful analysis of costs and benefits is needed to determine when MAS is advantageous compared to conventional breeding methods.