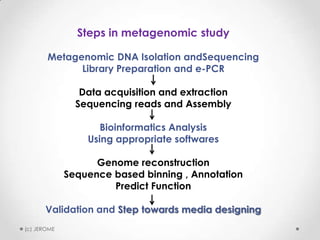
Metagenomics is the study of microbial communities directly in their natural environments without isolating individual species in the lab. It involves sequencing DNA from environmental samples and analyzing the metagenomes. Some key points are that metagenomics can identify uncultivable microbes, bypassing the need for culture, and it has led to advances in understanding microbial ecology, evolution, and diversity. The rumen, home to a complex microbial community important for ruminant digestion, is a important target of metagenomics study. Next generation sequencing techniques now allow more accessible exploration of microbial systems through metagenomics.