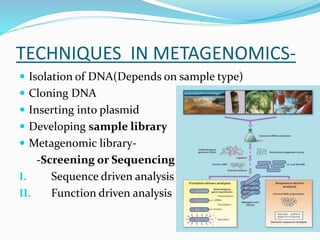
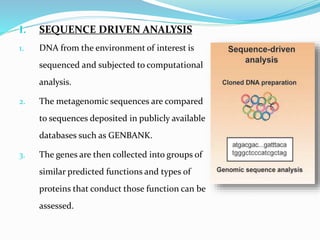
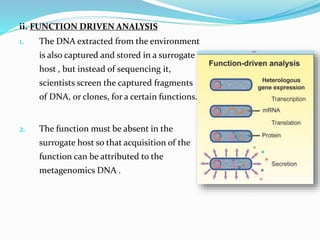
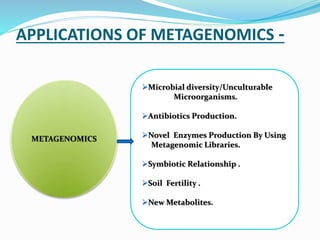
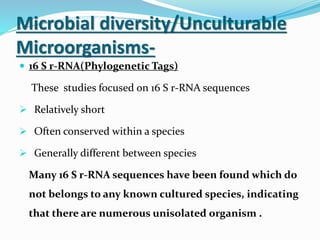
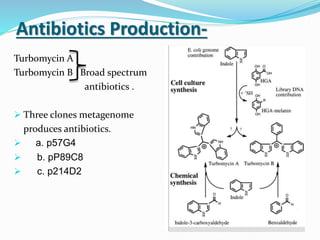
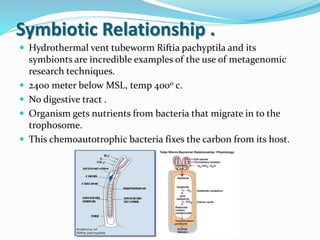
This document discusses metagenomics, which is the study of genetic material recovered directly from environmental samples without culturing organisms. It outlines the difference between traditional genomics which studies one organism at a time in culture, versus metagenomics which sequences all DNA in a sample without culturing. The document then covers historical events in metagenomics, techniques used including direct DNA extraction and sequencing or function-based screening, applications such as discovering microbial diversity and novel enzymes, and future directions such as understanding human microbiomes and discovering novel pathways and organisms.