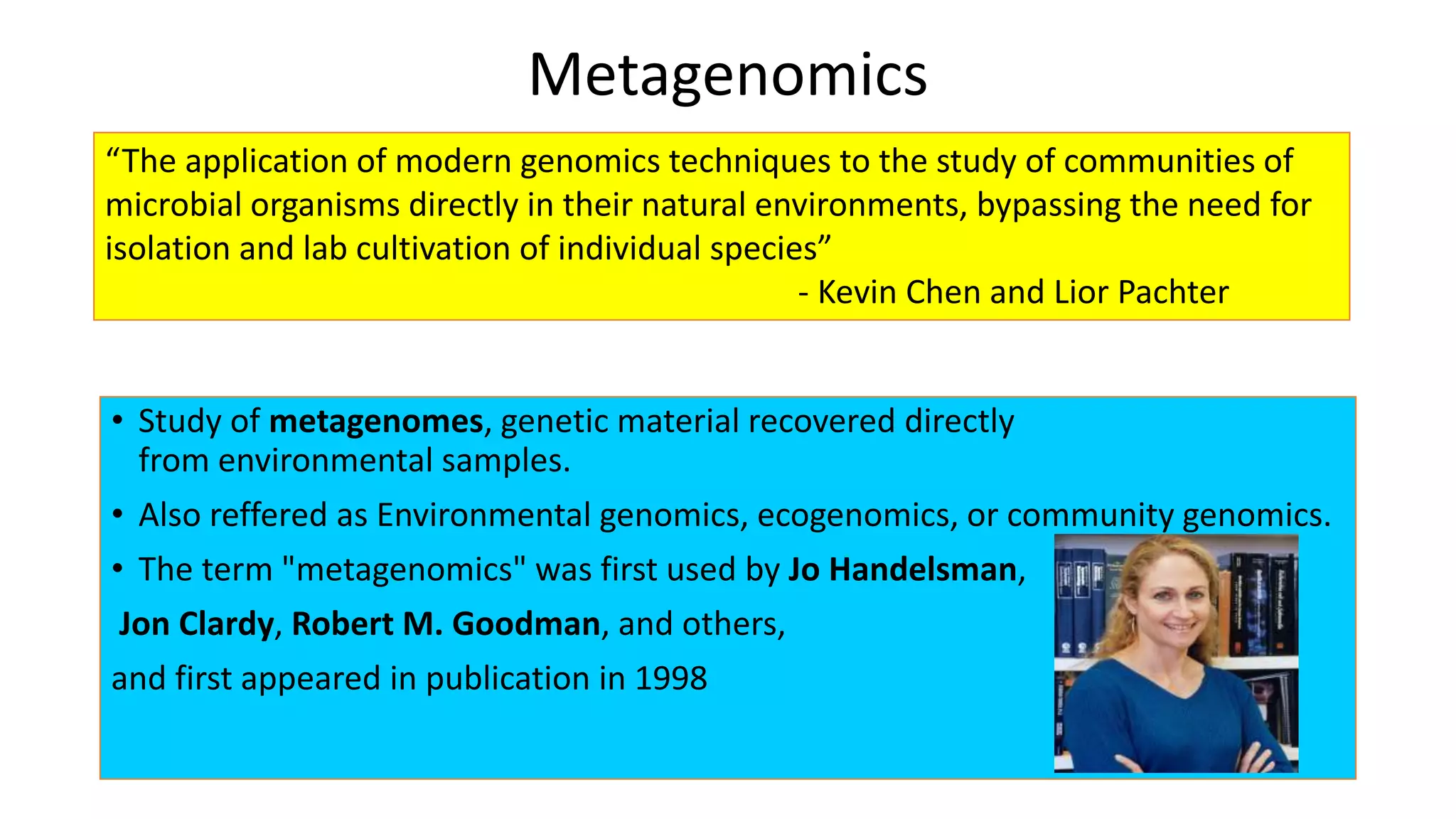
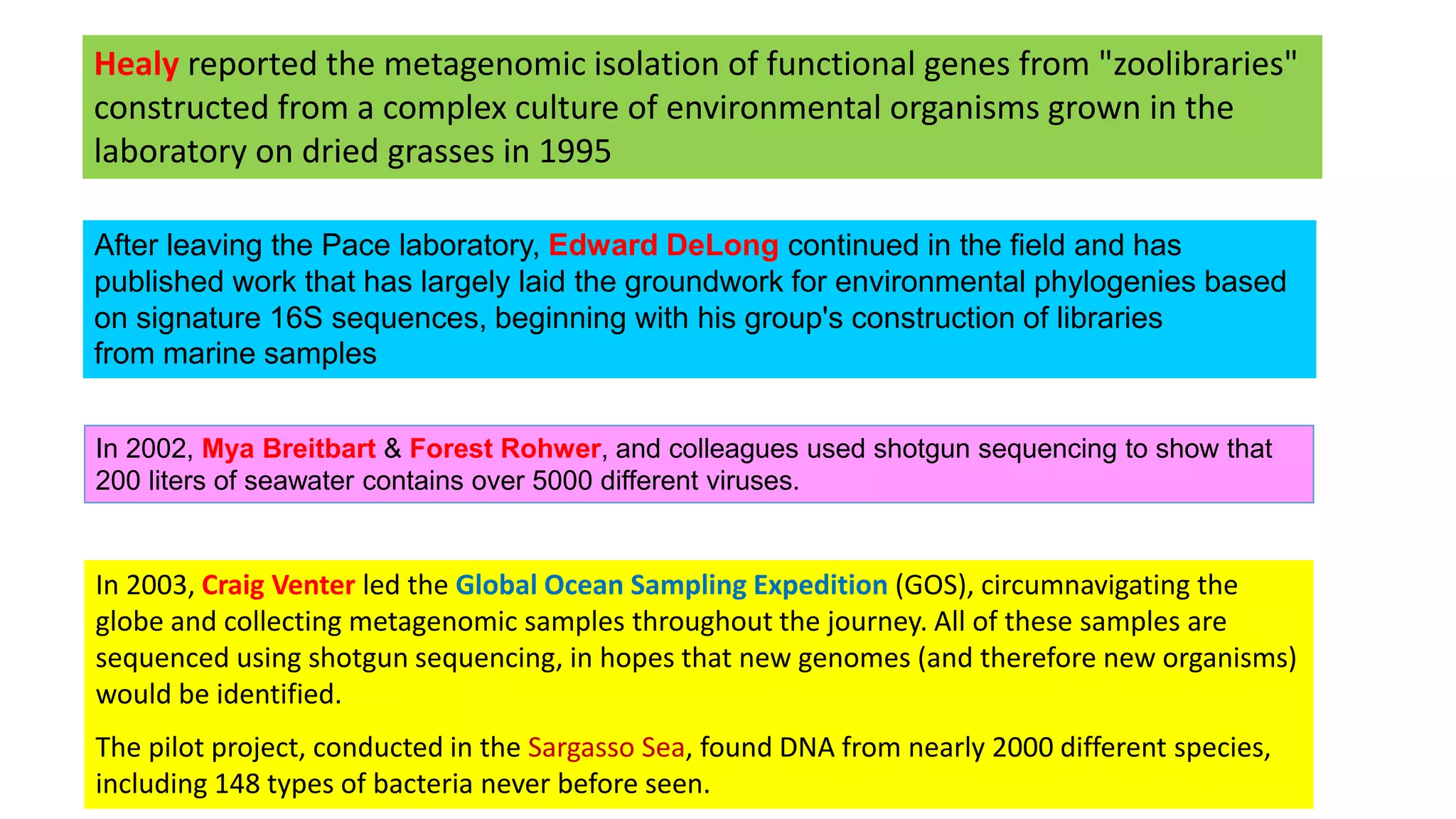
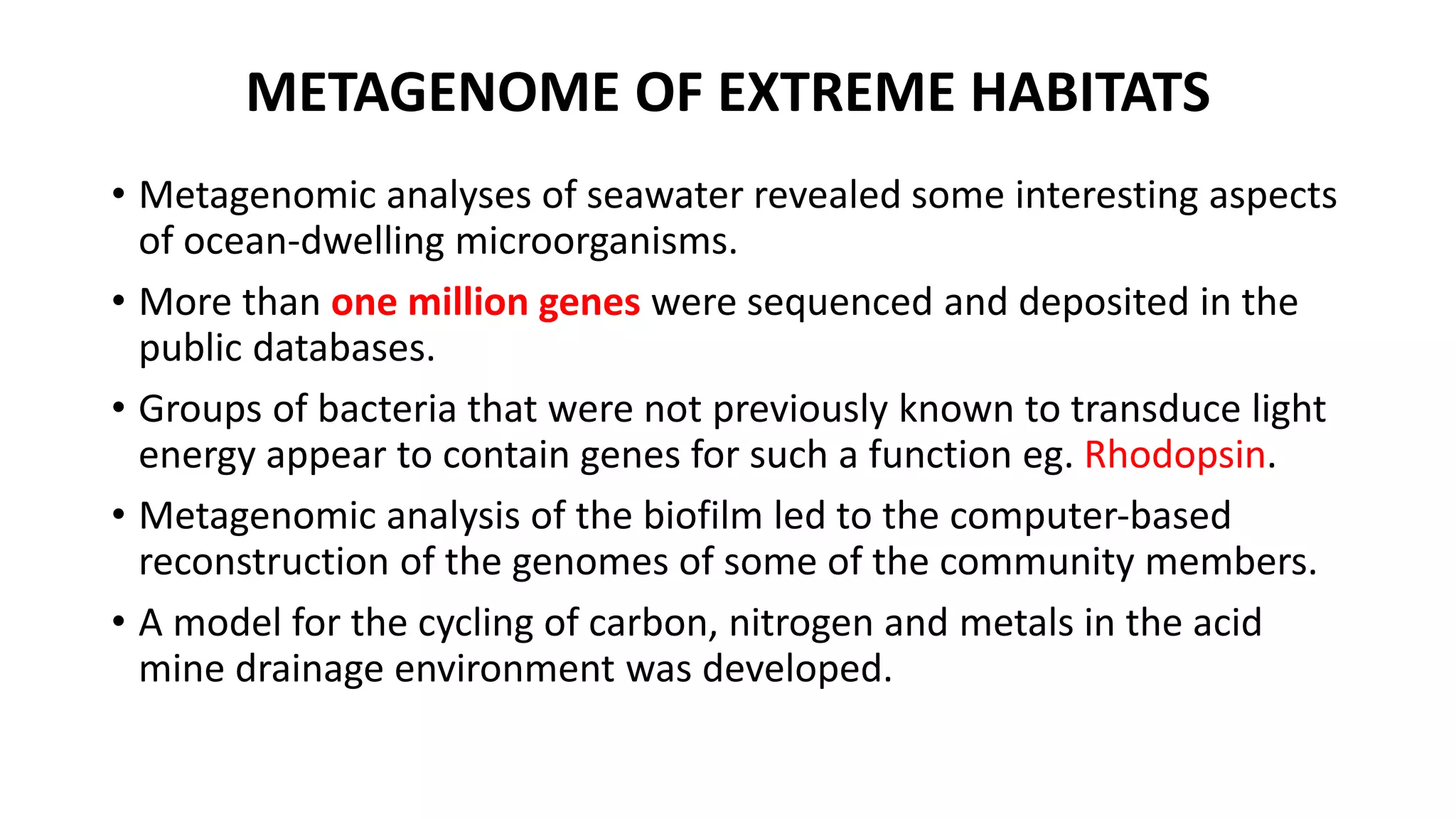
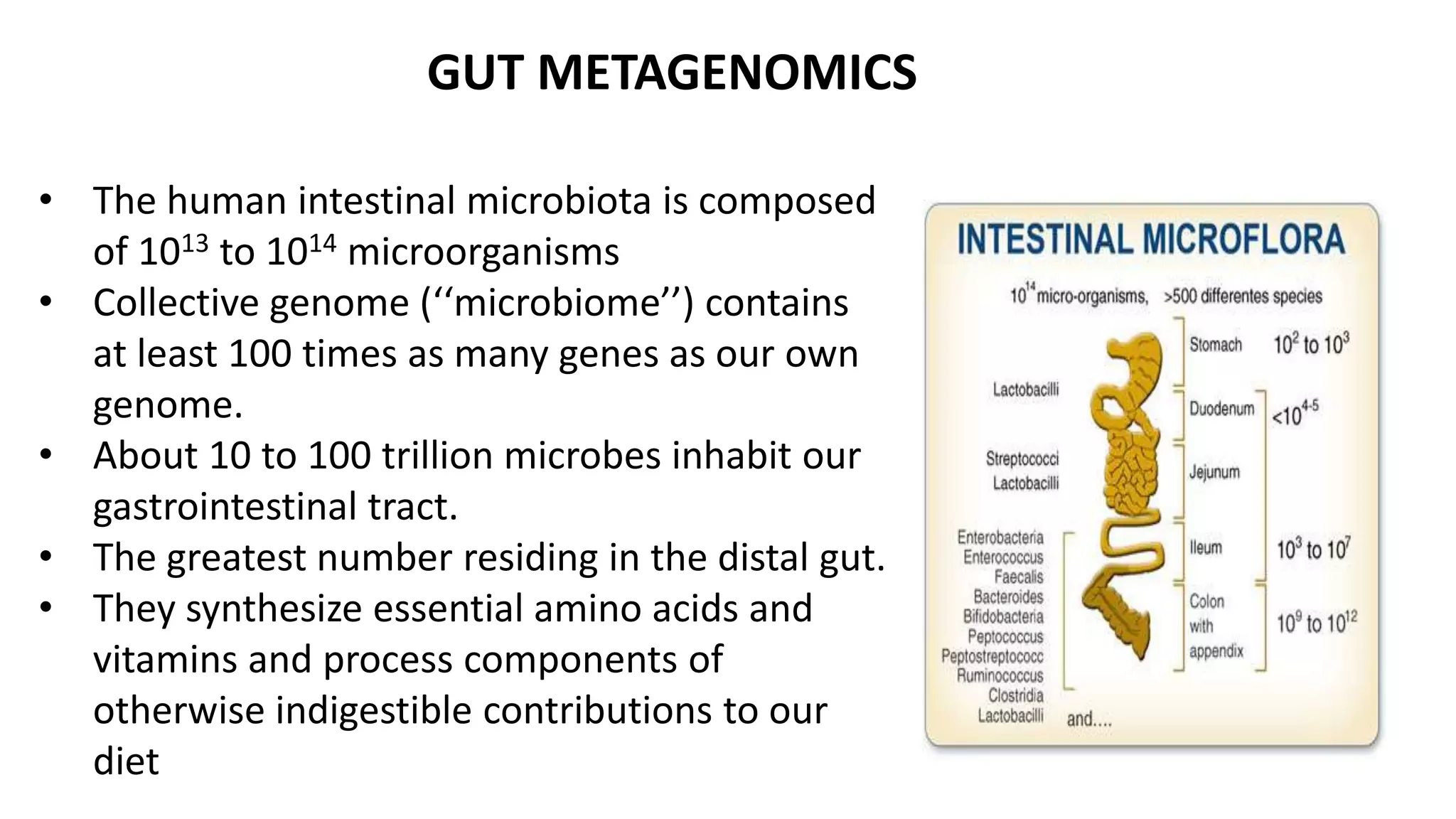
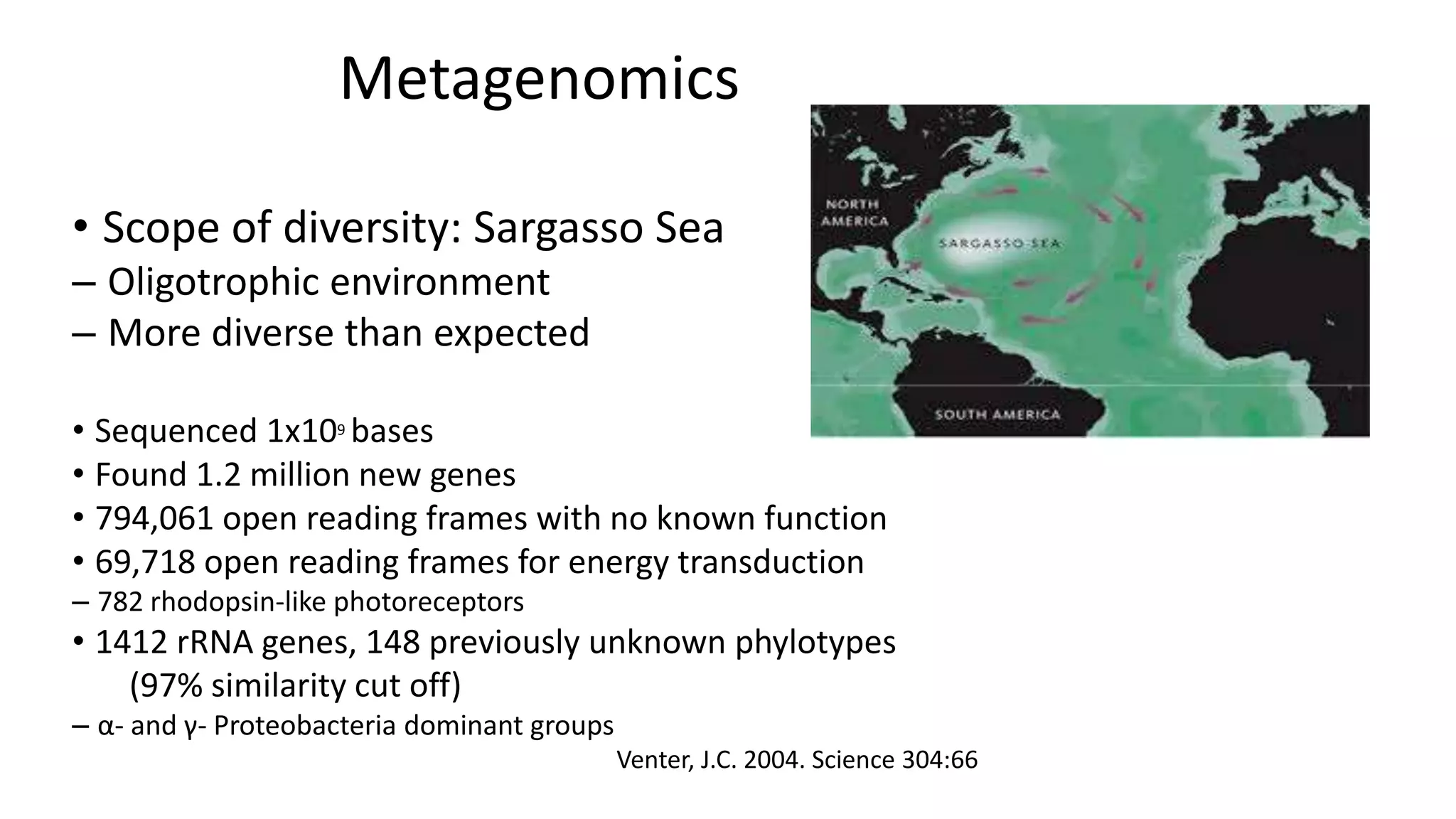
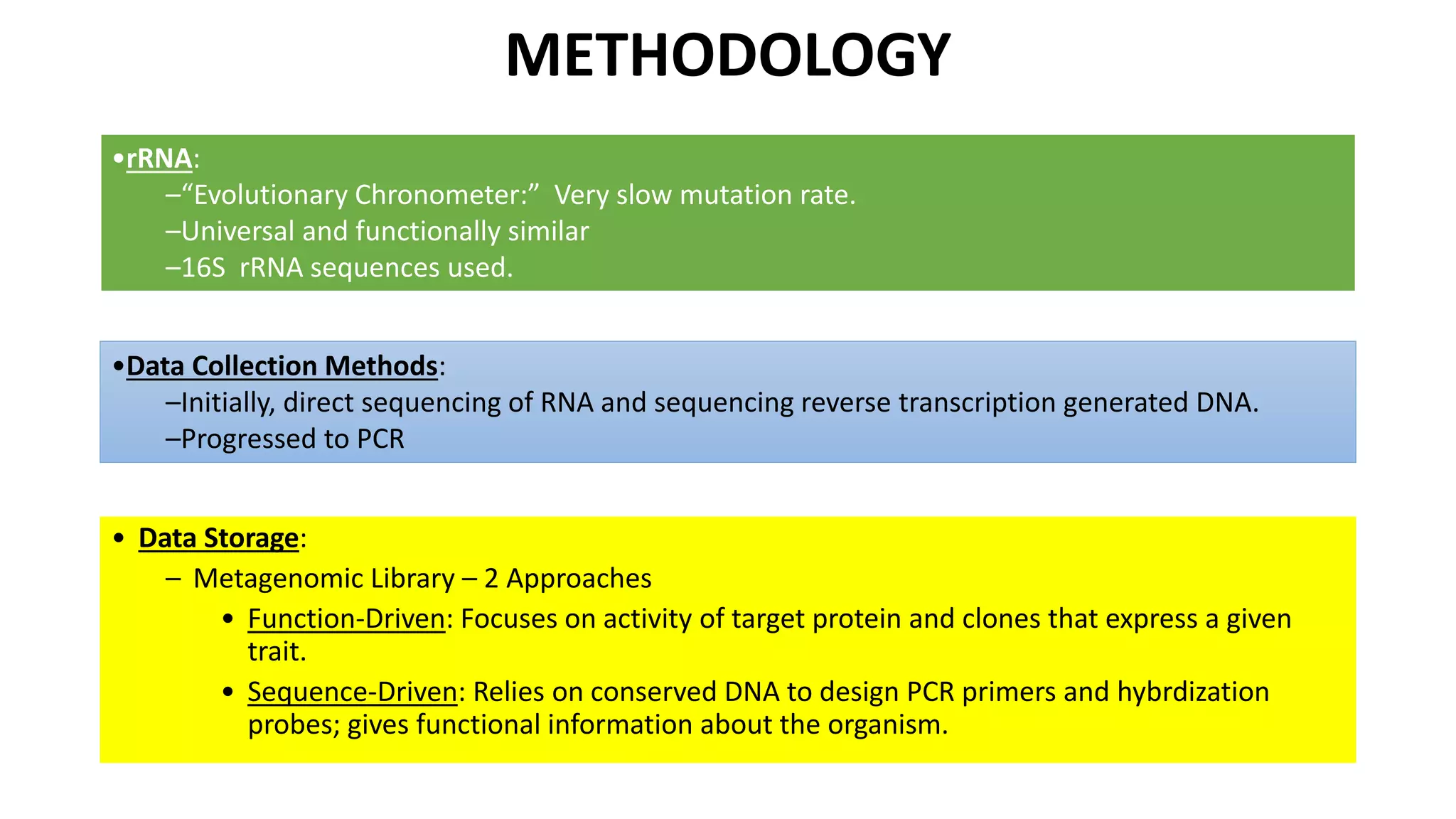
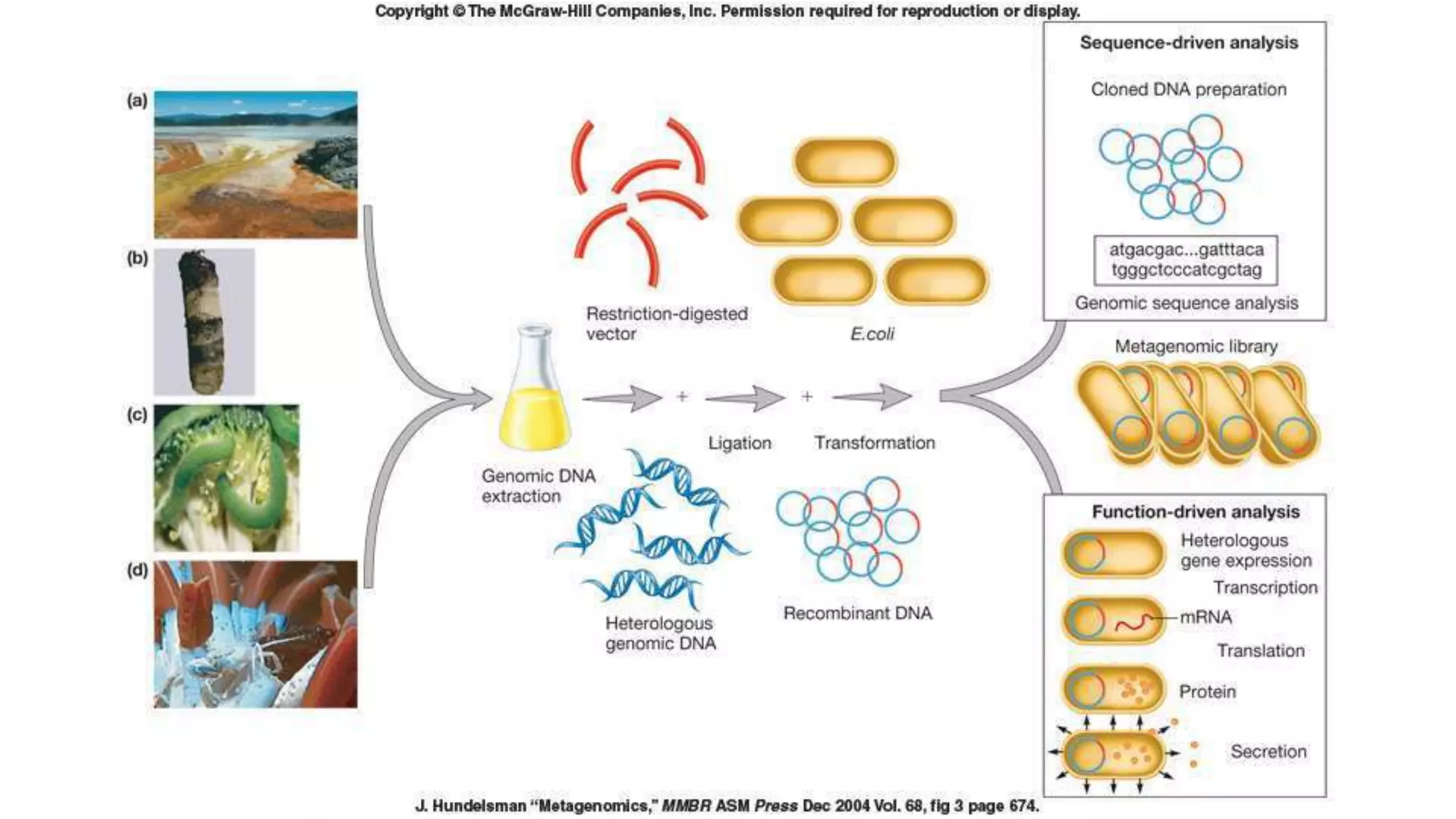
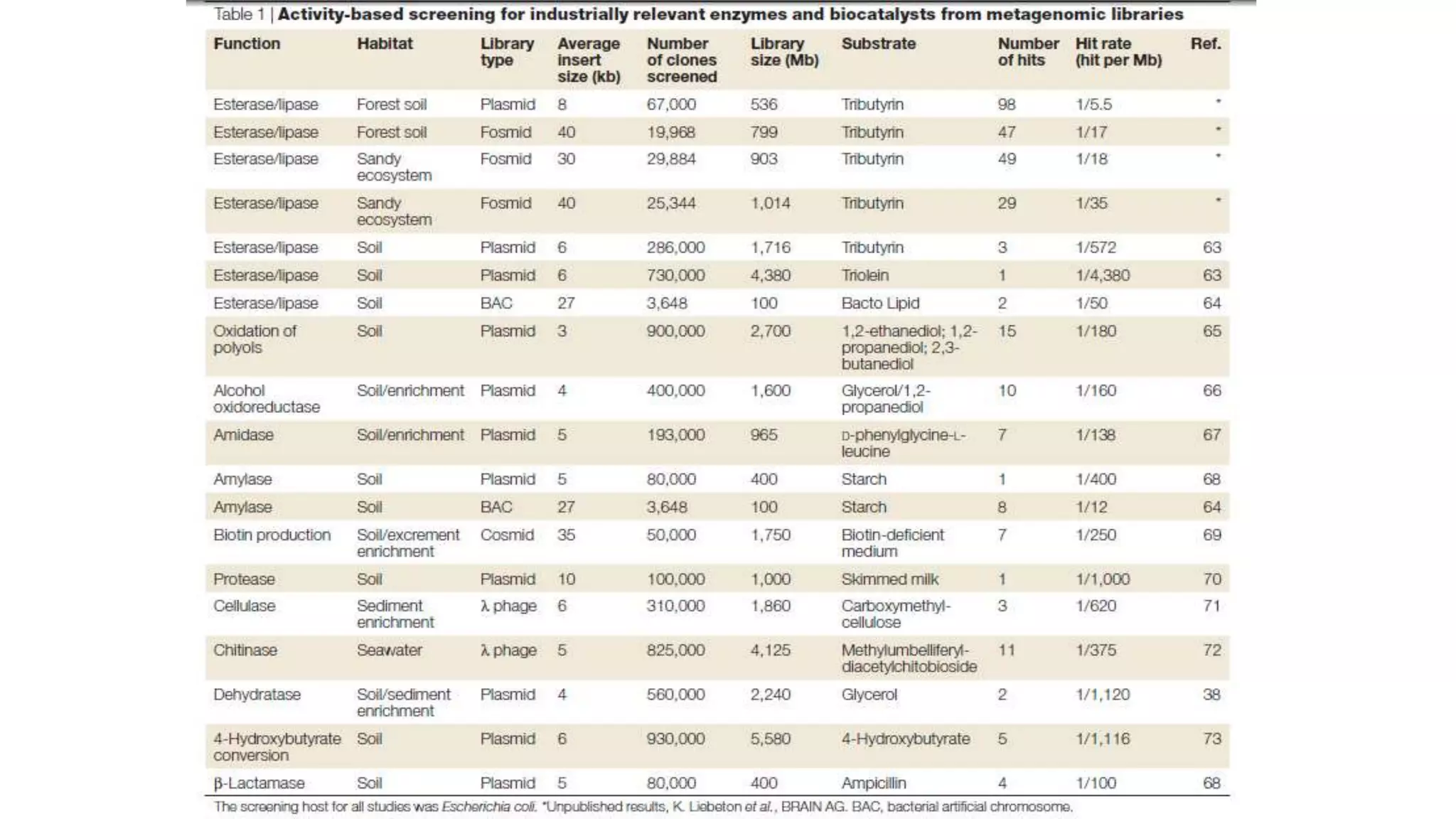
This document discusses the field of metagenomics, which involves directly extracting and sequencing genetic material from environmental samples without culturing individual microbial species. It provides a brief history of metagenomics from early microbiologists in the 17th century to recent large-scale sequencing projects. Methods of metagenomic analysis like sequence-driven and function-driven approaches are described. Applications to studying uncultured symbiotic microbes, extreme environments, and the human gut microbiome are also summarized.