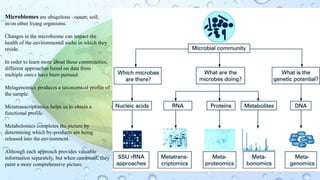
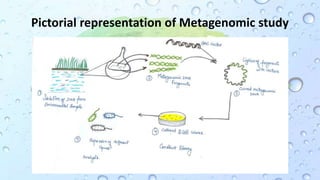
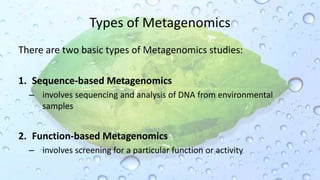
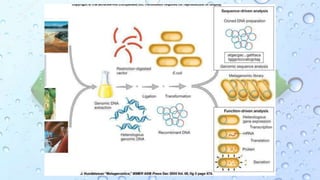
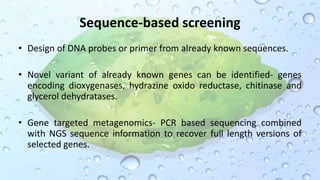
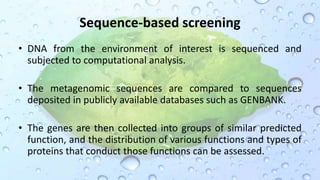
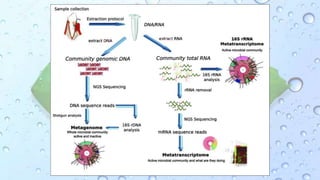
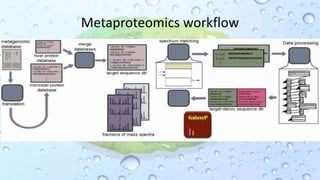
Microbiomes are found in various environments, and their changes can affect local health; thus, diverse omics approaches like metagenomics, metatranscriptomics, and metabolomics are utilized to study these communities. Metagenomics allows for functional and sequence-based analysis of microbes without isolation and identifies biological diversity, while metatranscriptomics examines transcribed sequences to understand metabolic capacity, and metaproteomics analyzes proteins to link genetic diversity with ecosystem functions. Despite their limitations, combining these methods offers deeper insights into microbial communities and their ecological impacts.