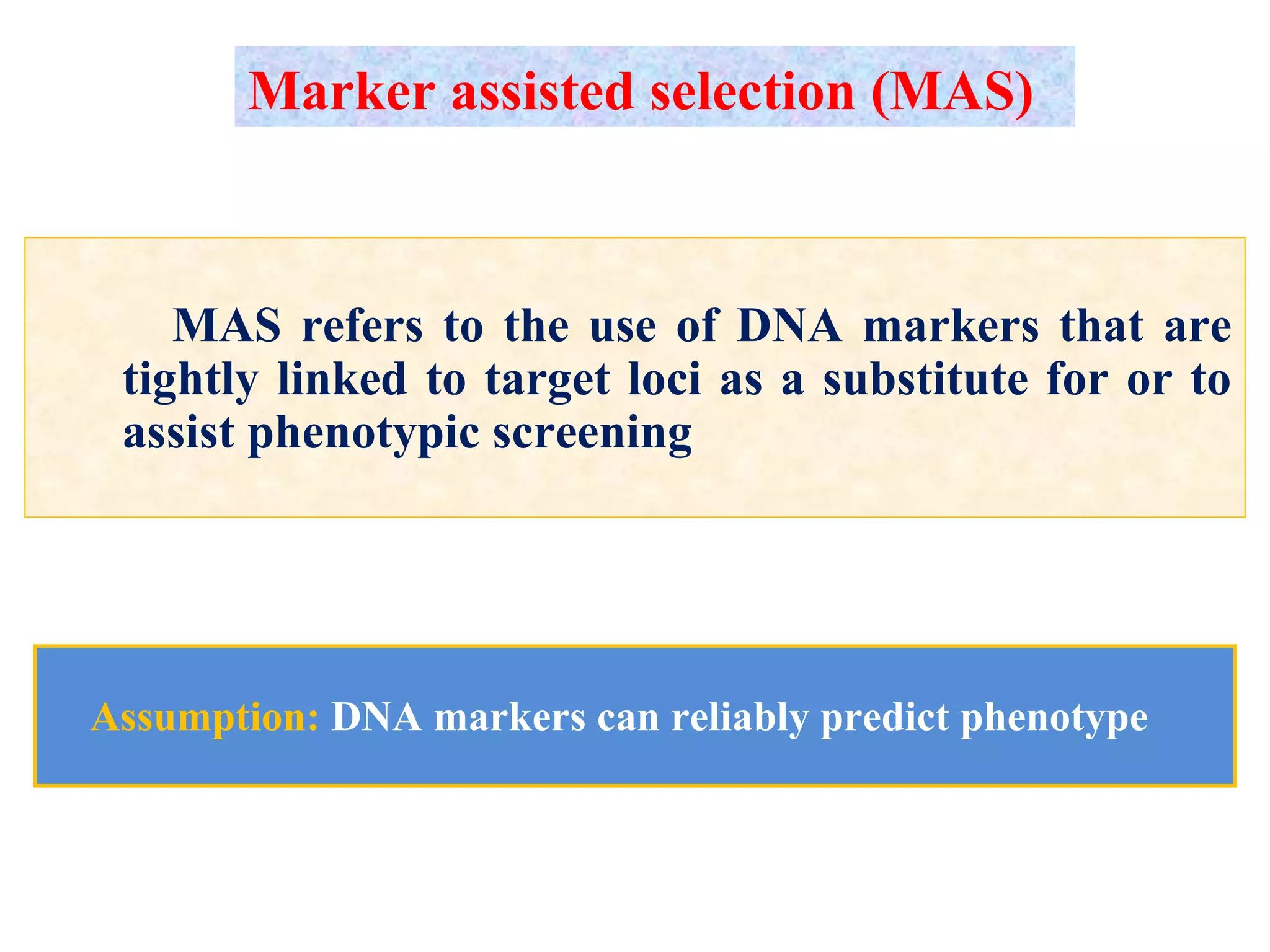
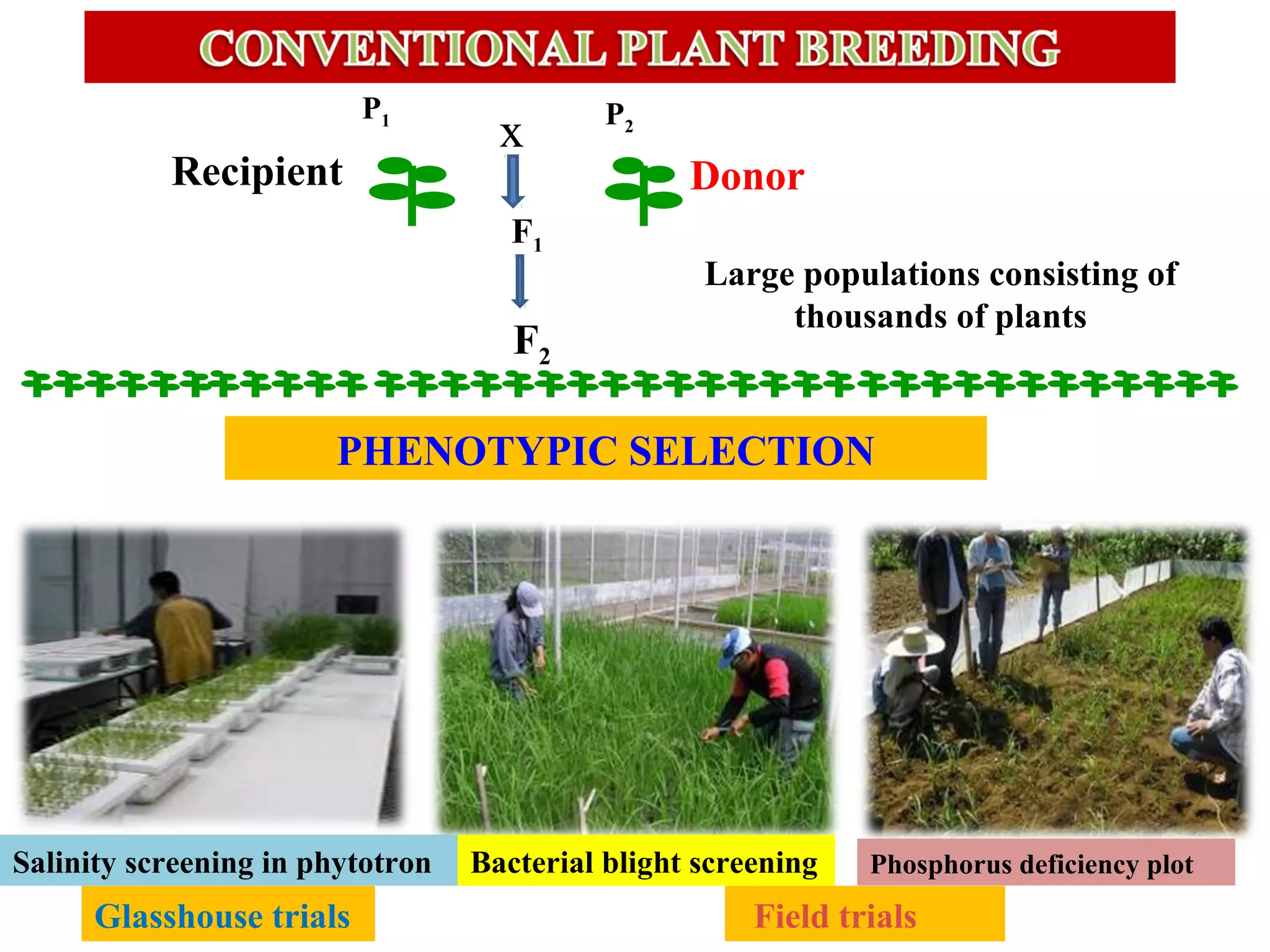
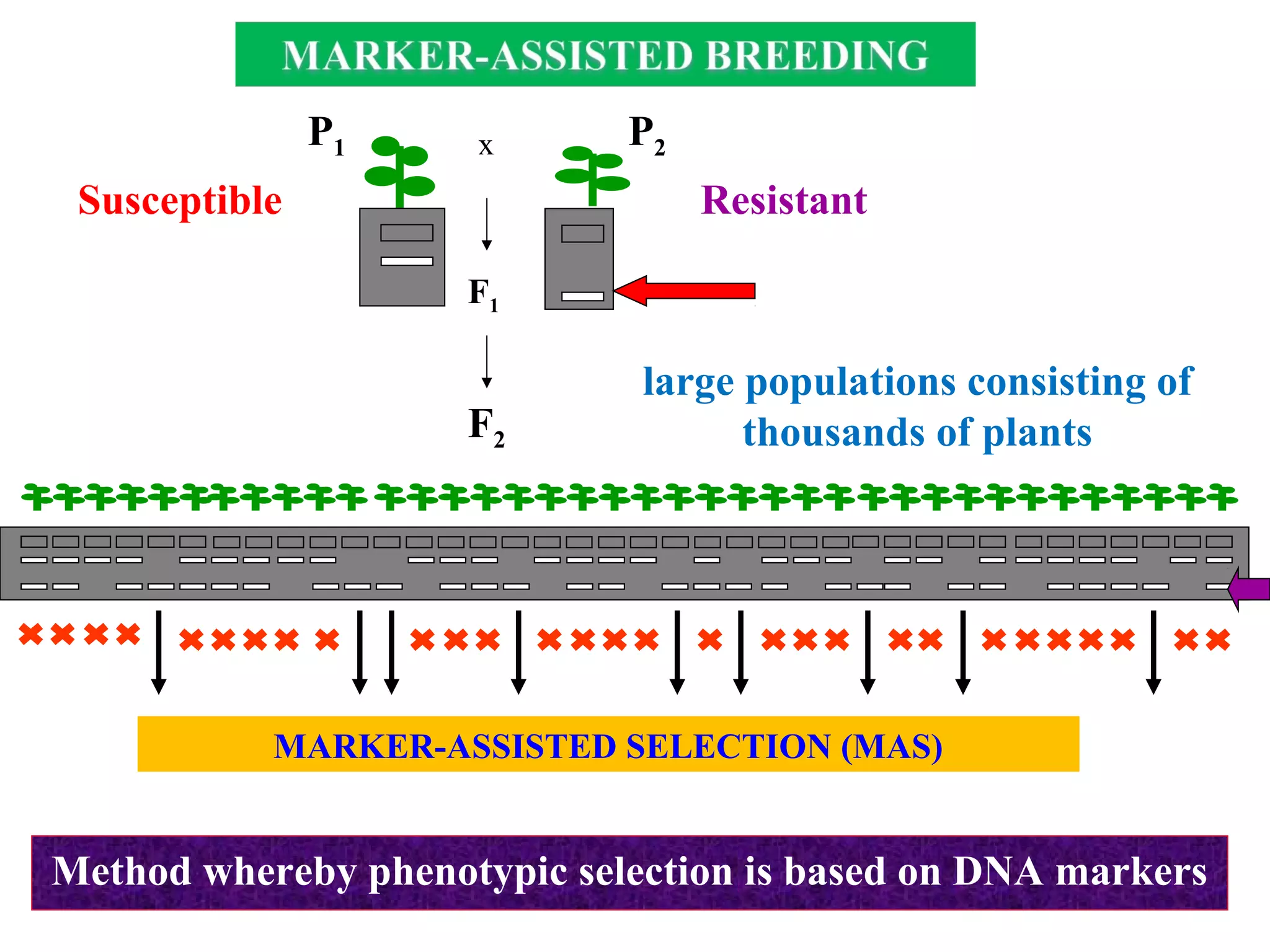
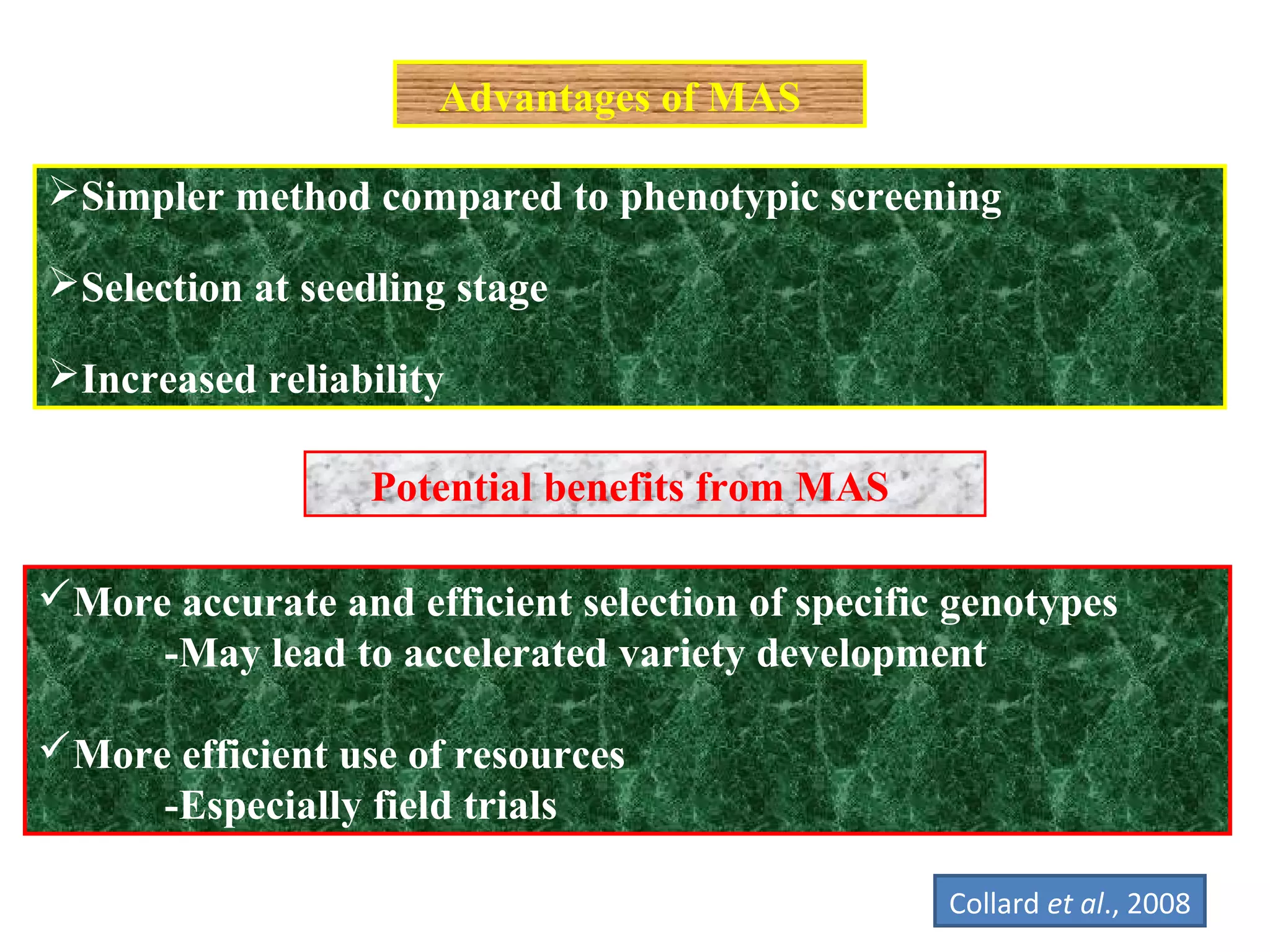
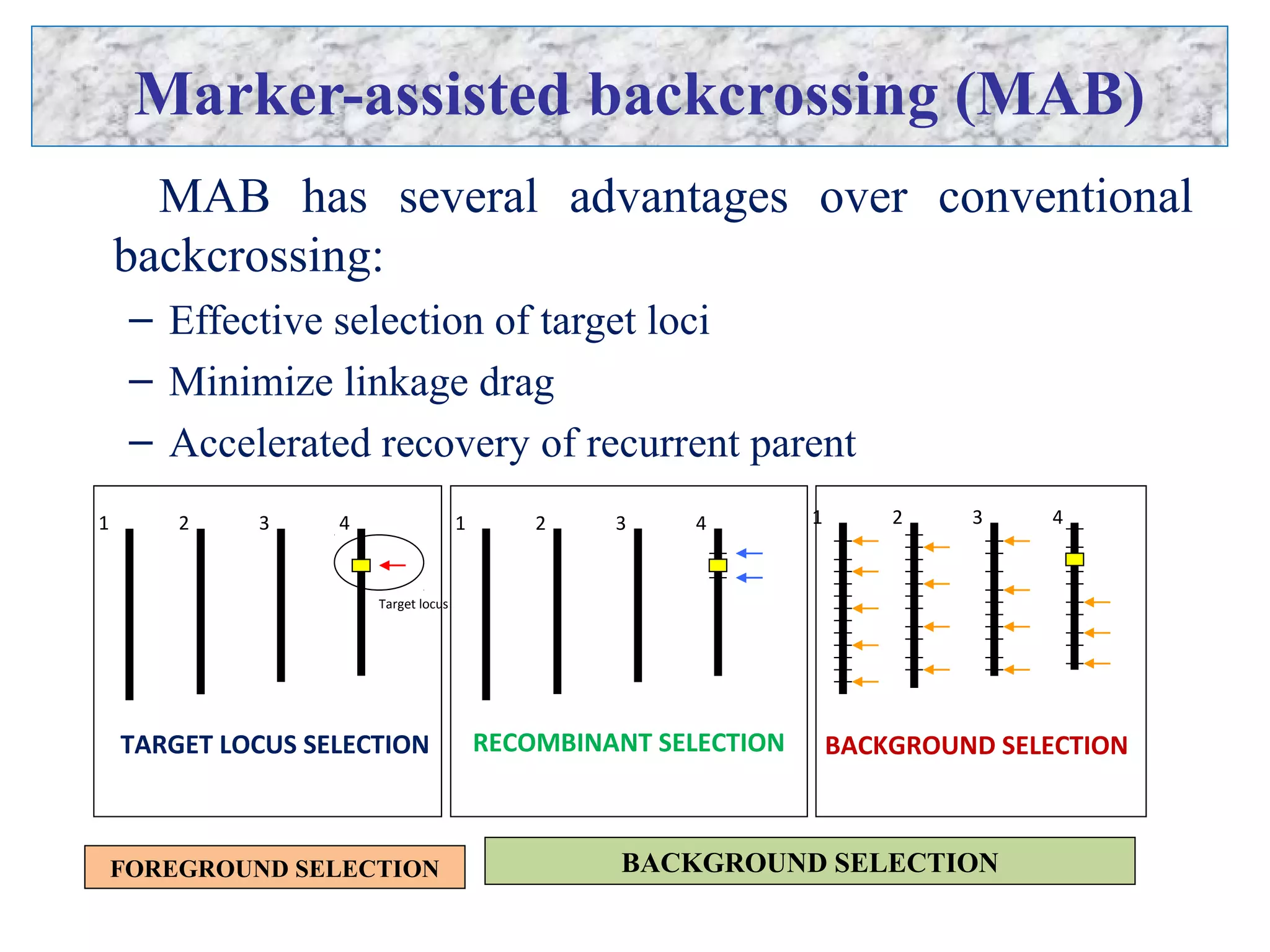
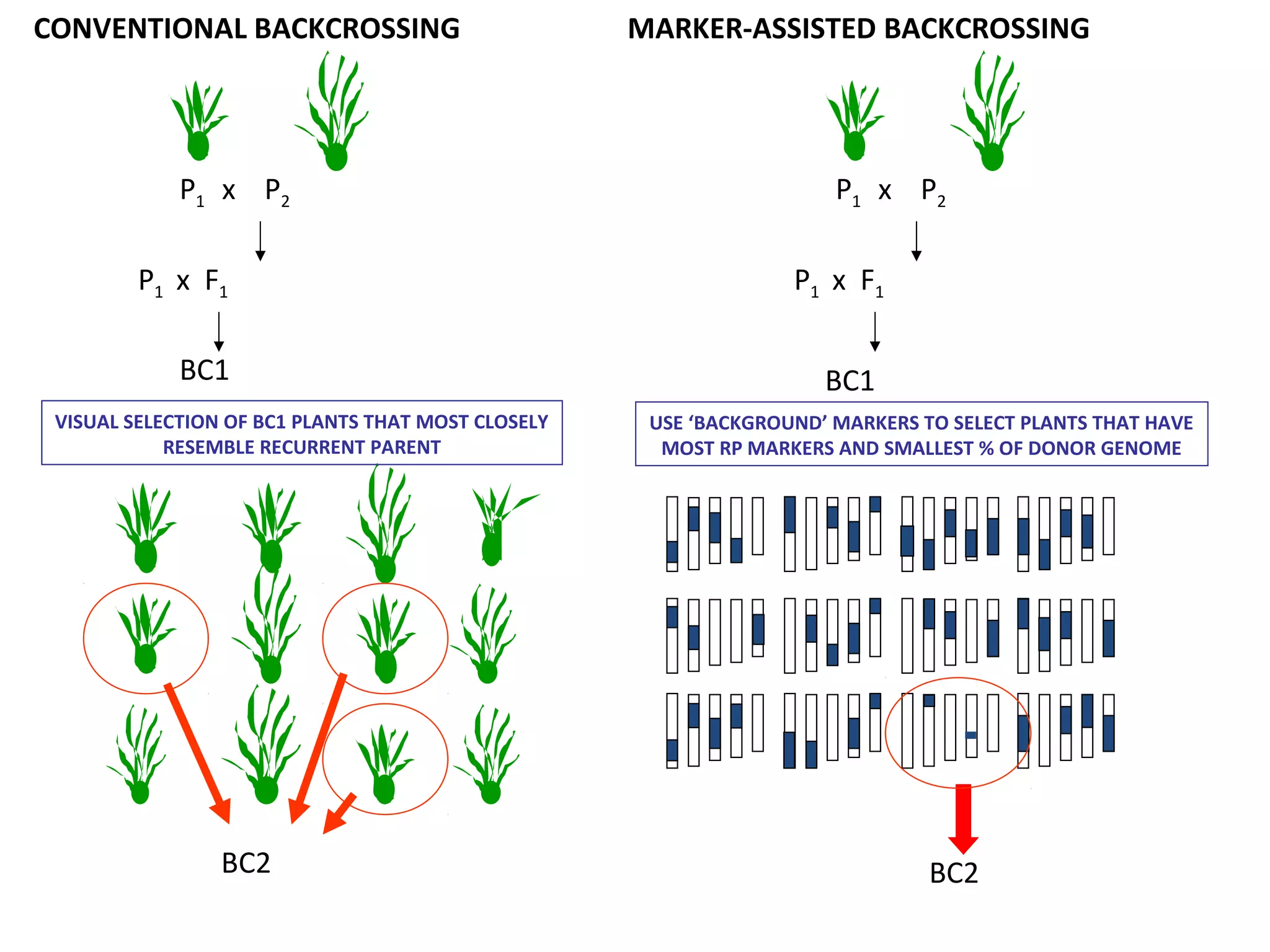
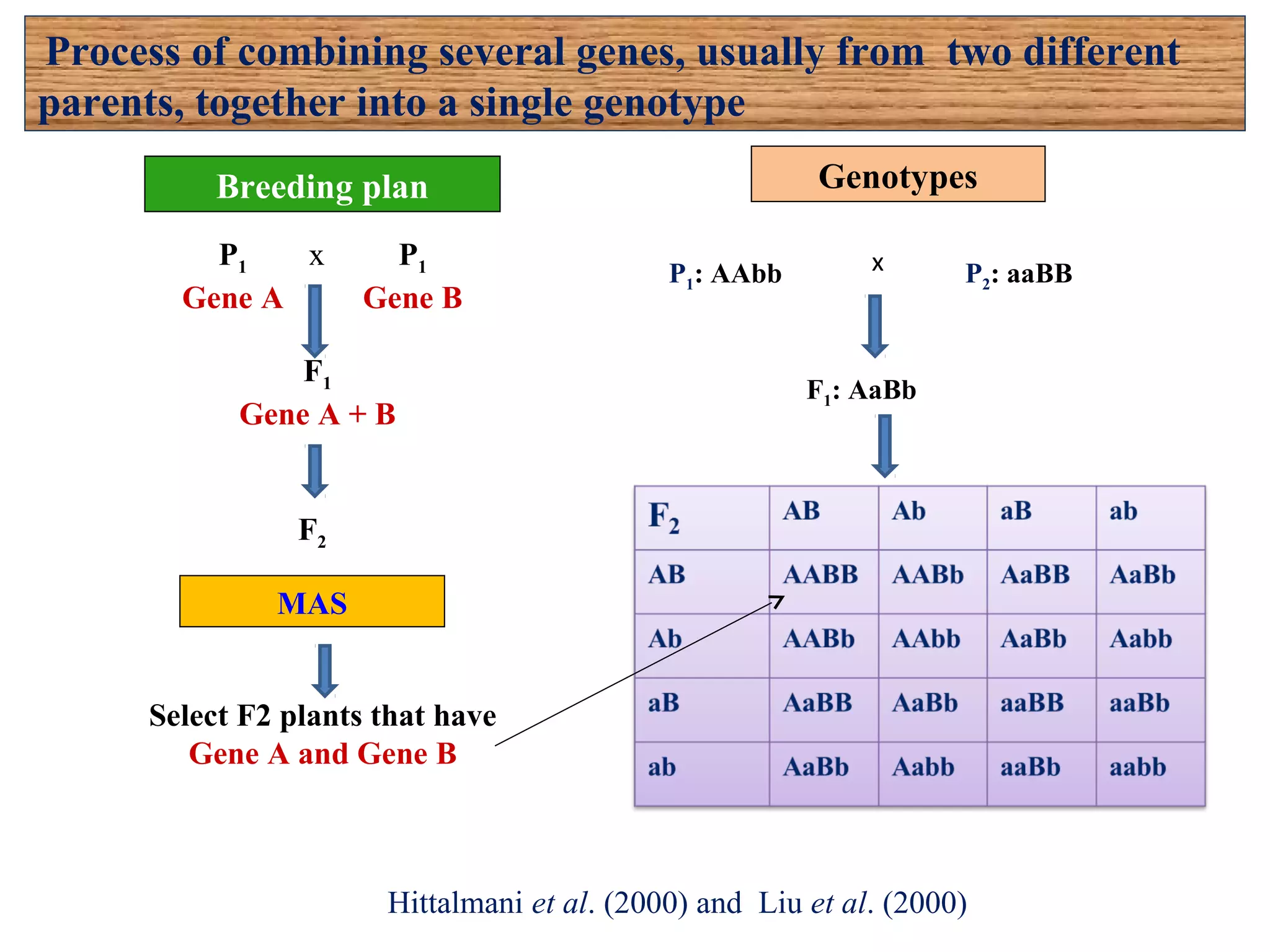
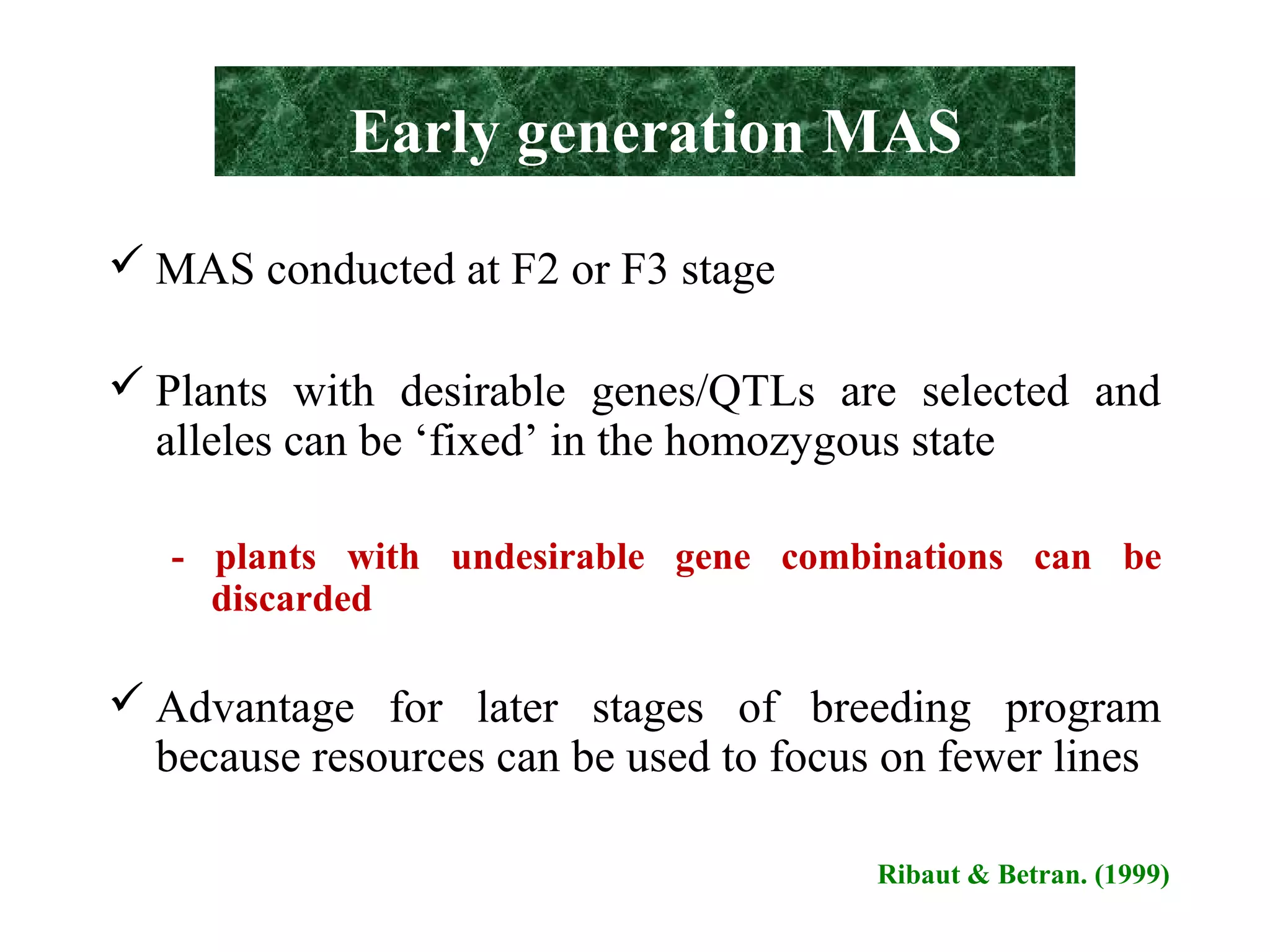
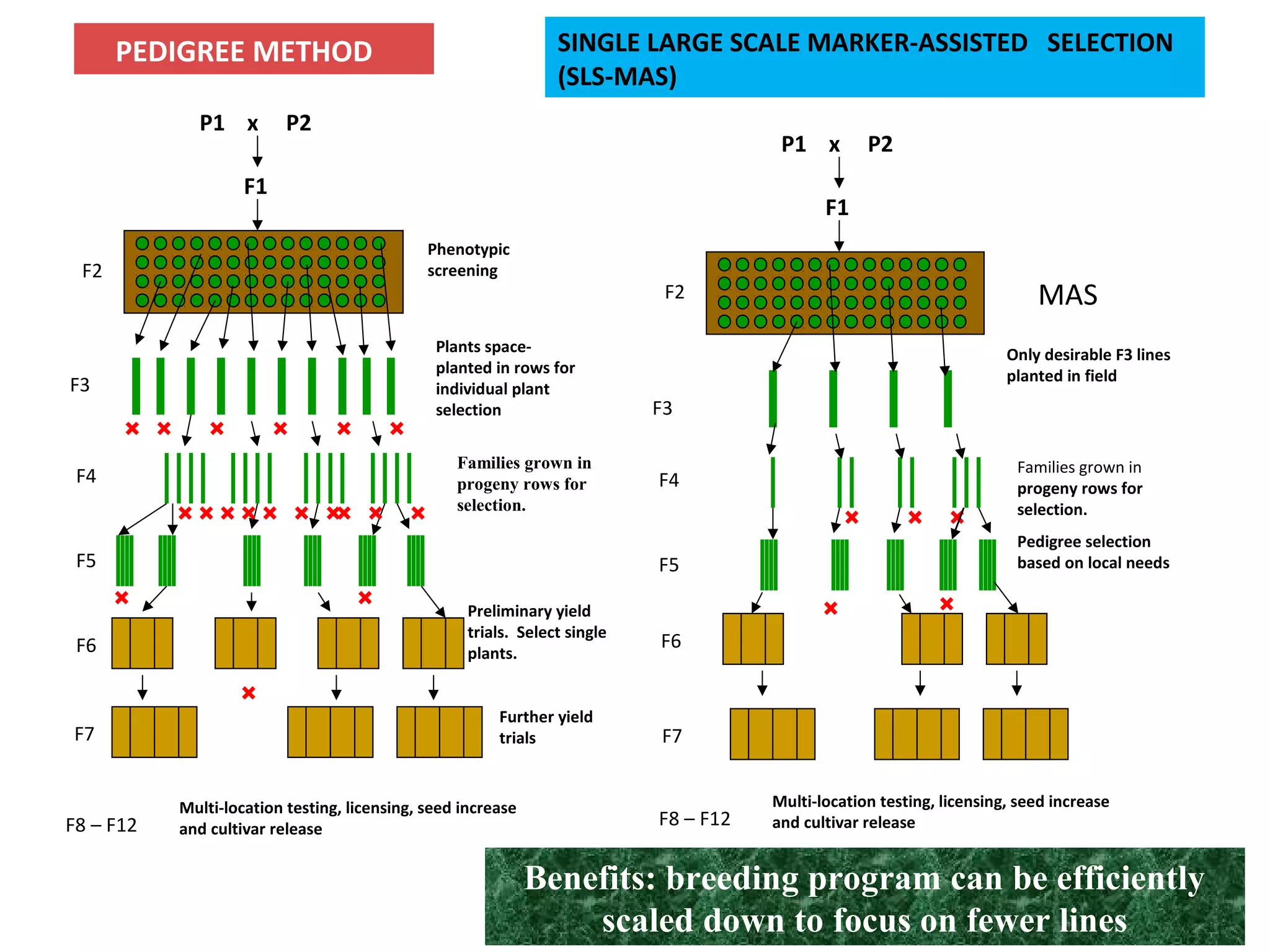
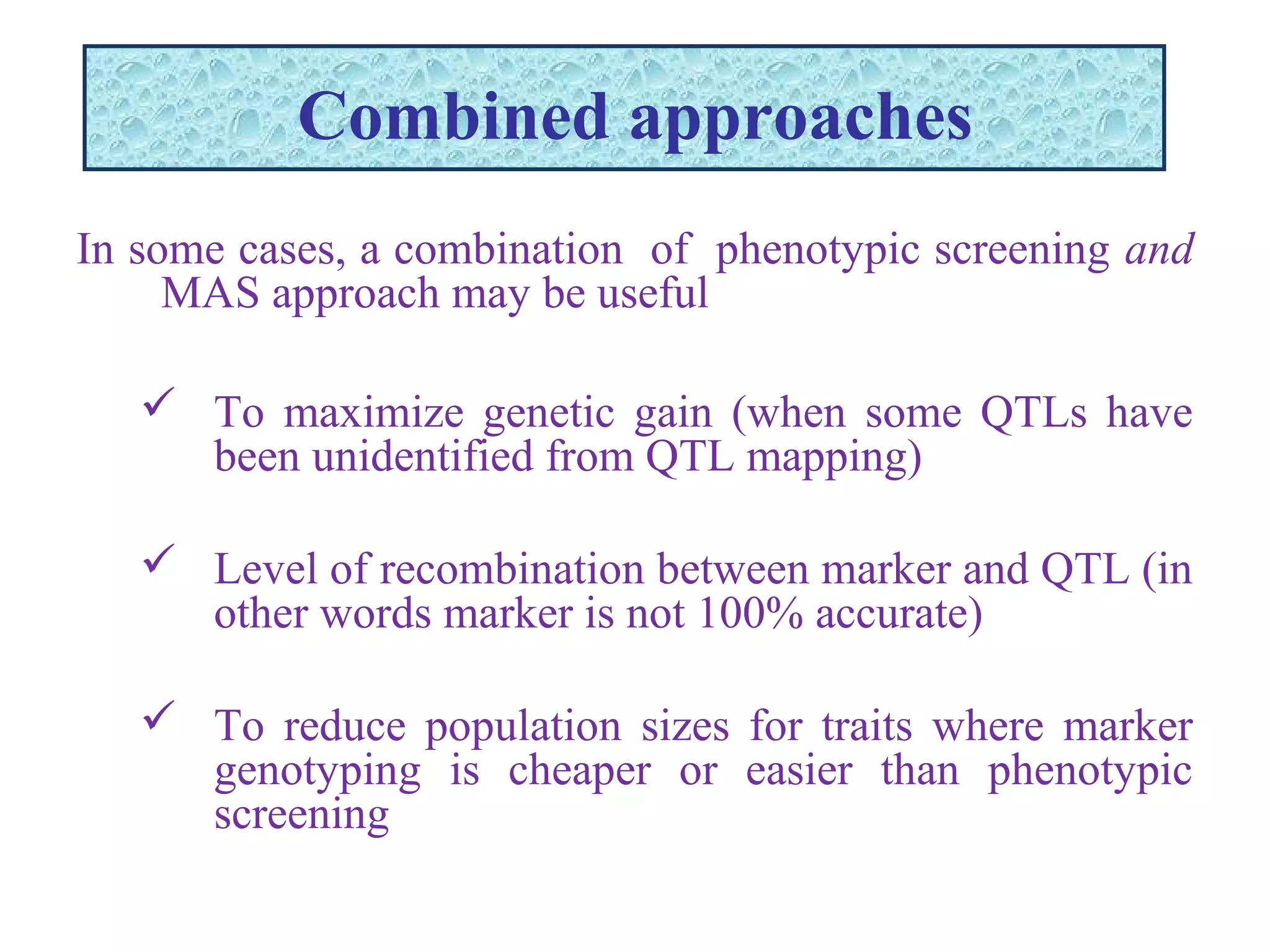
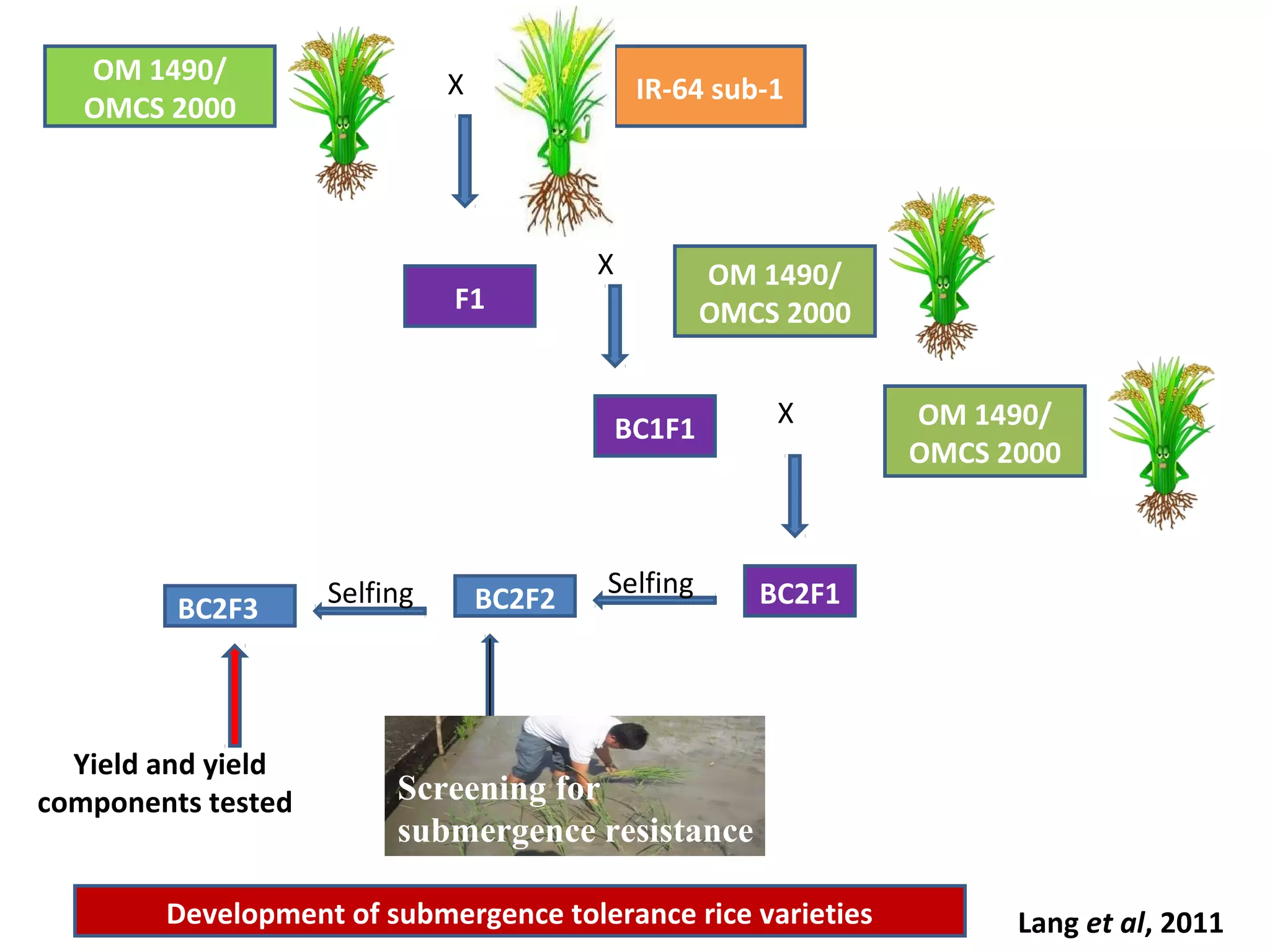
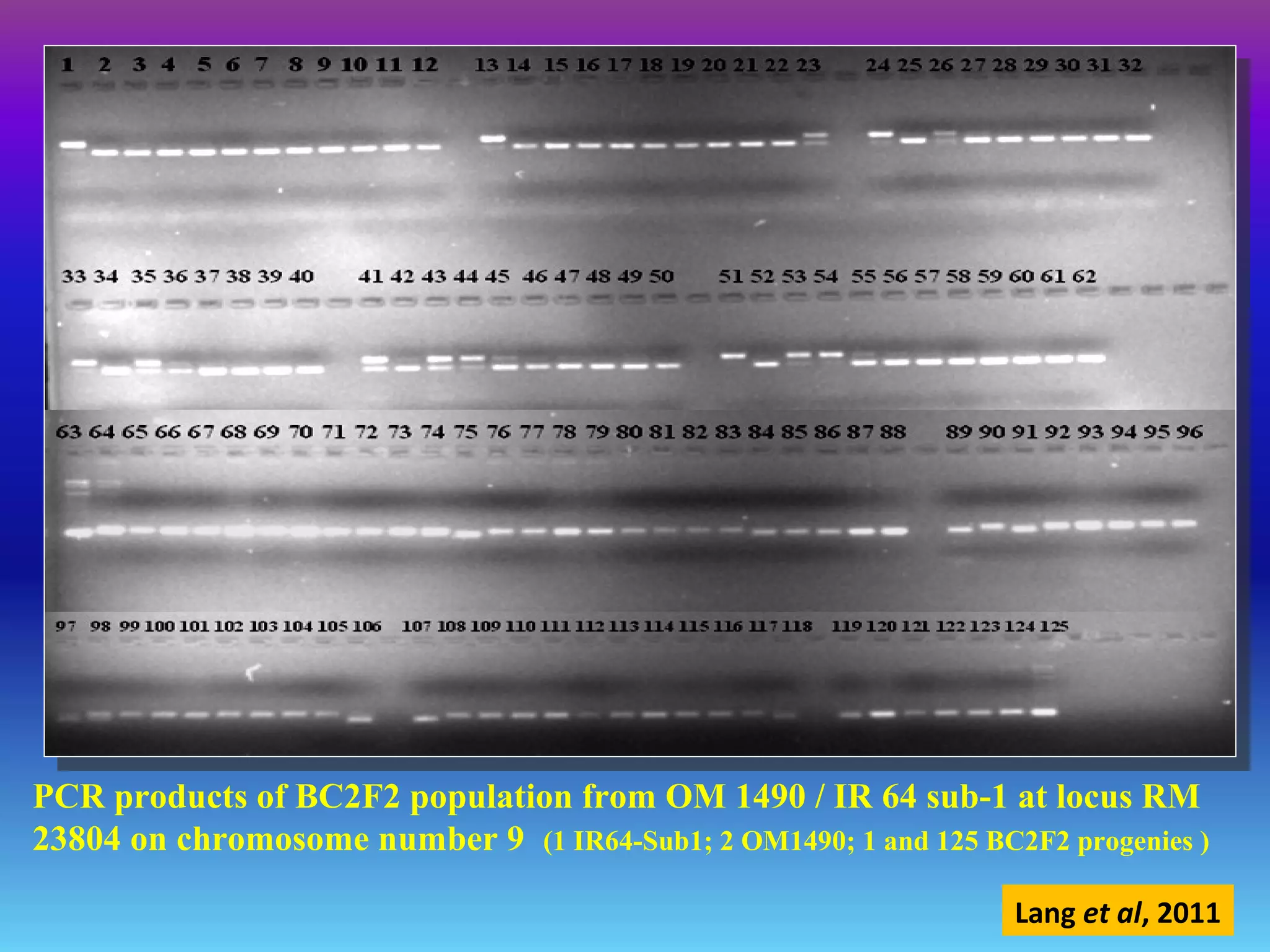
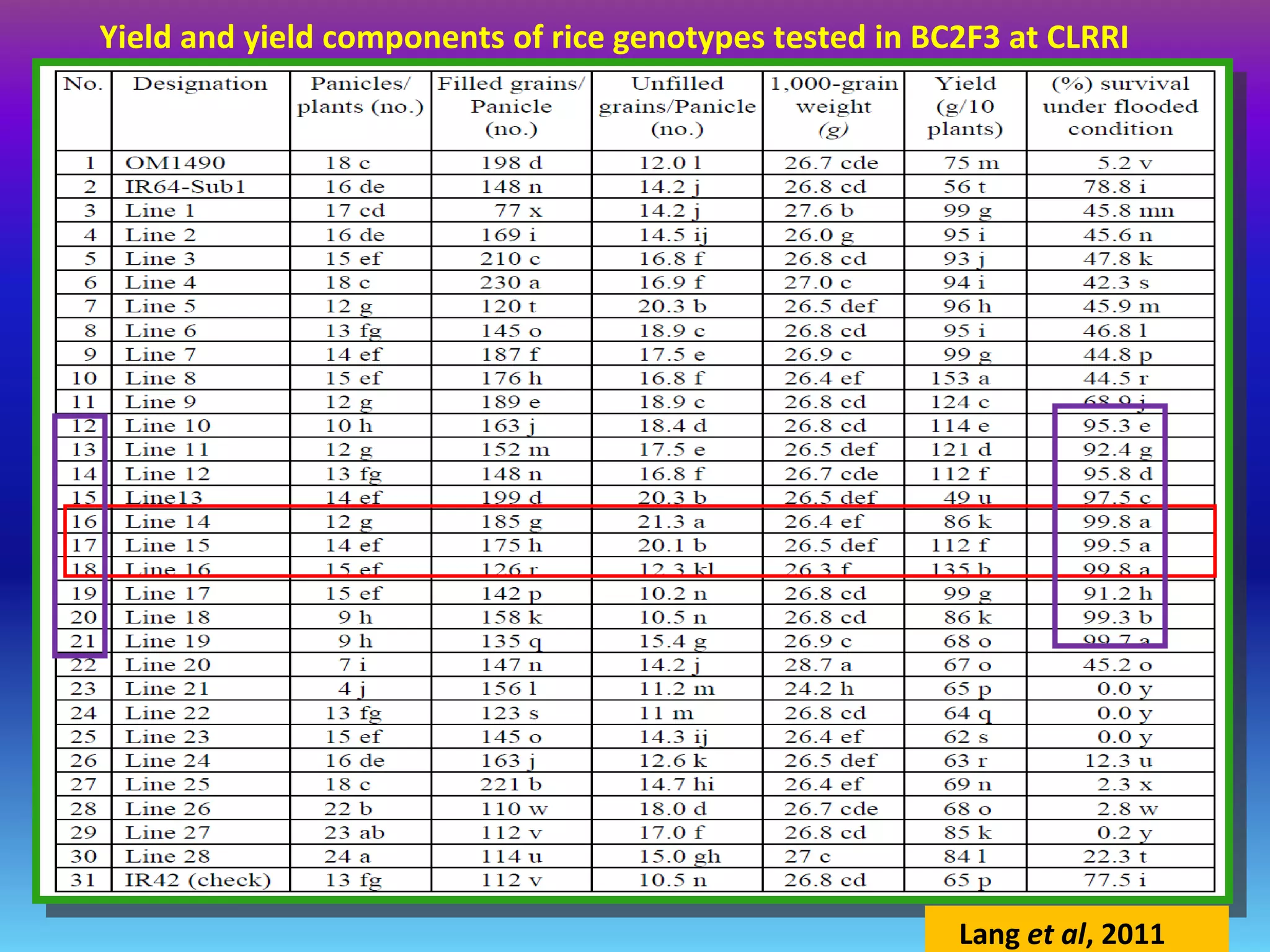
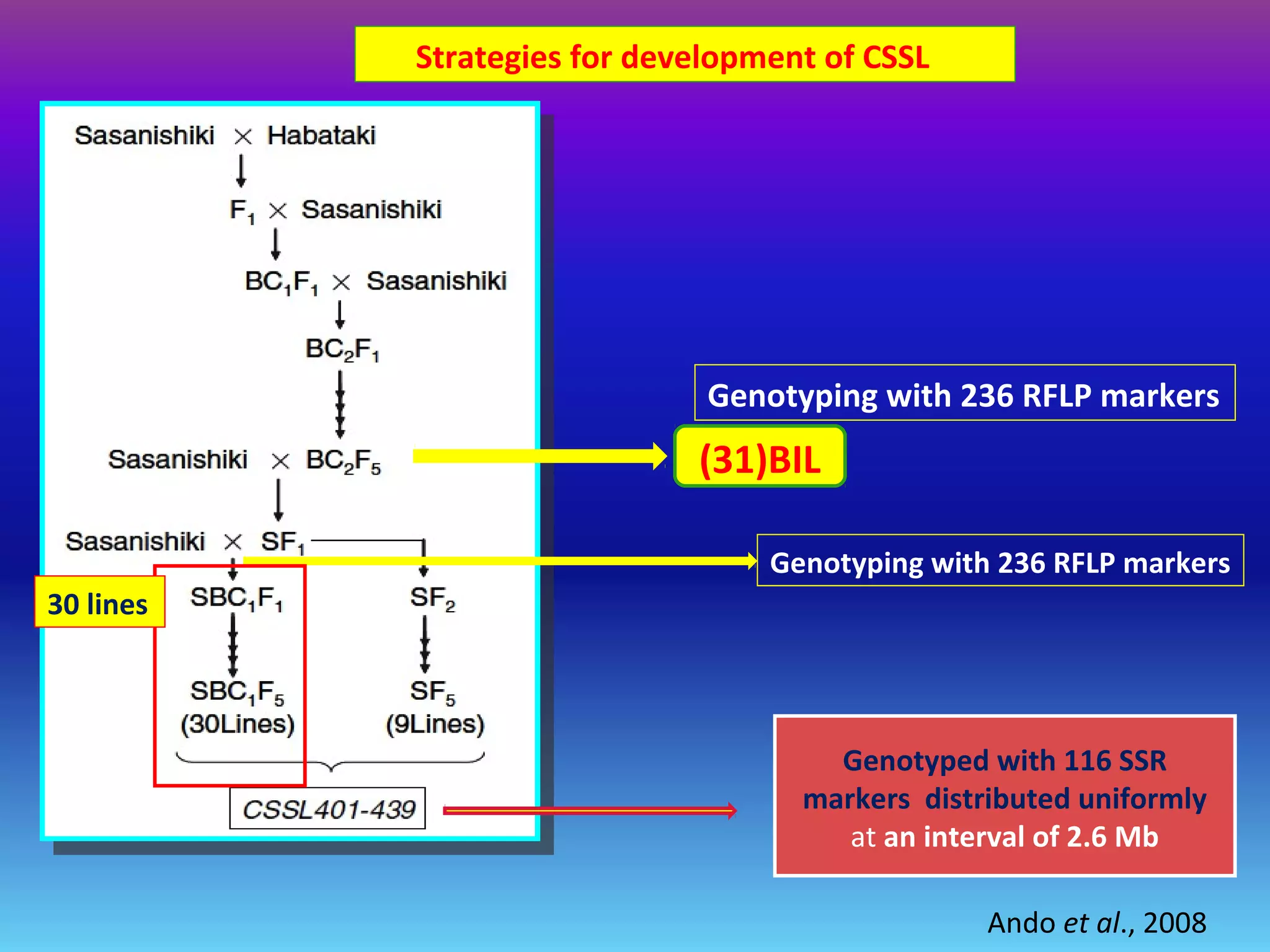
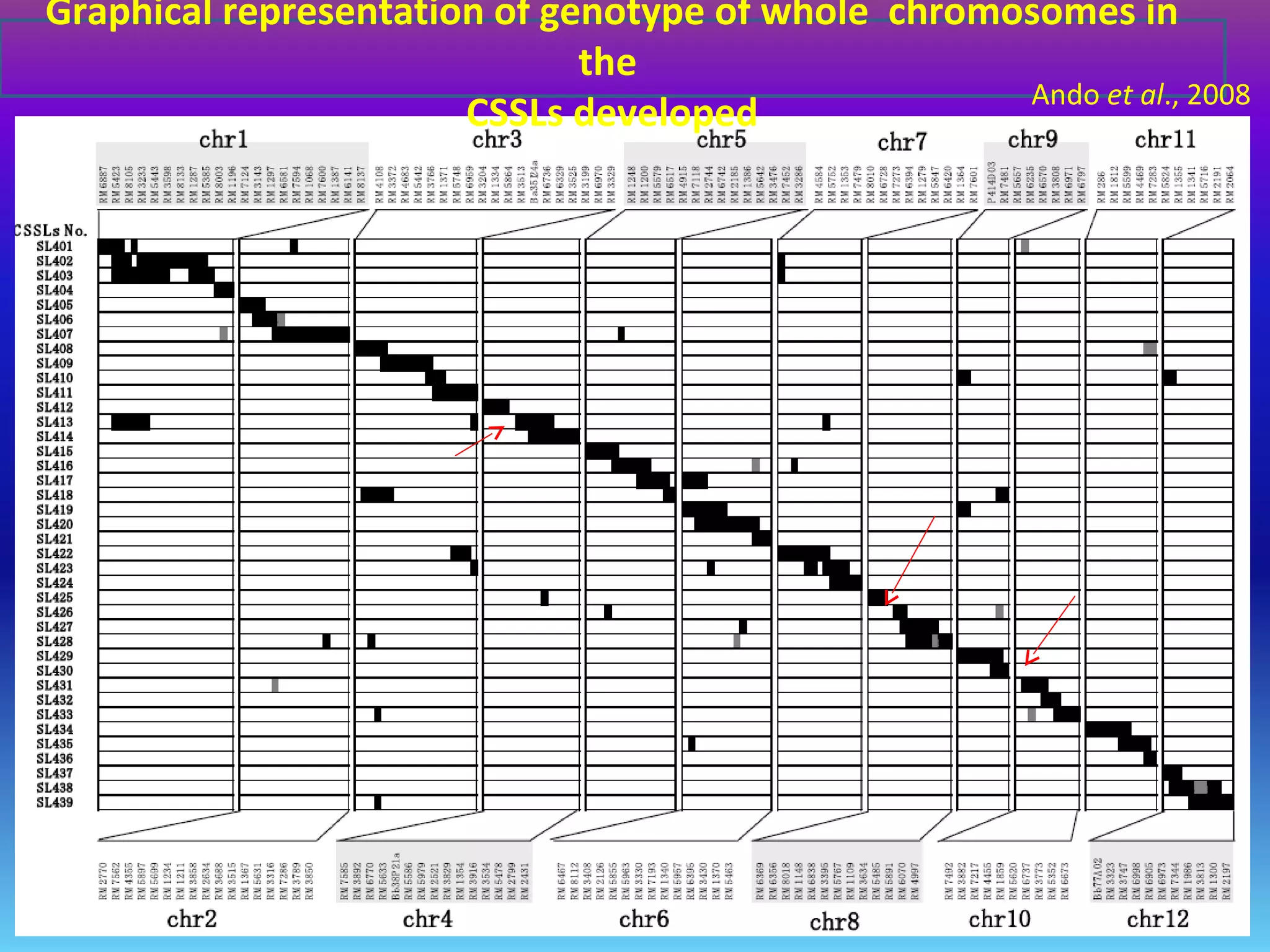
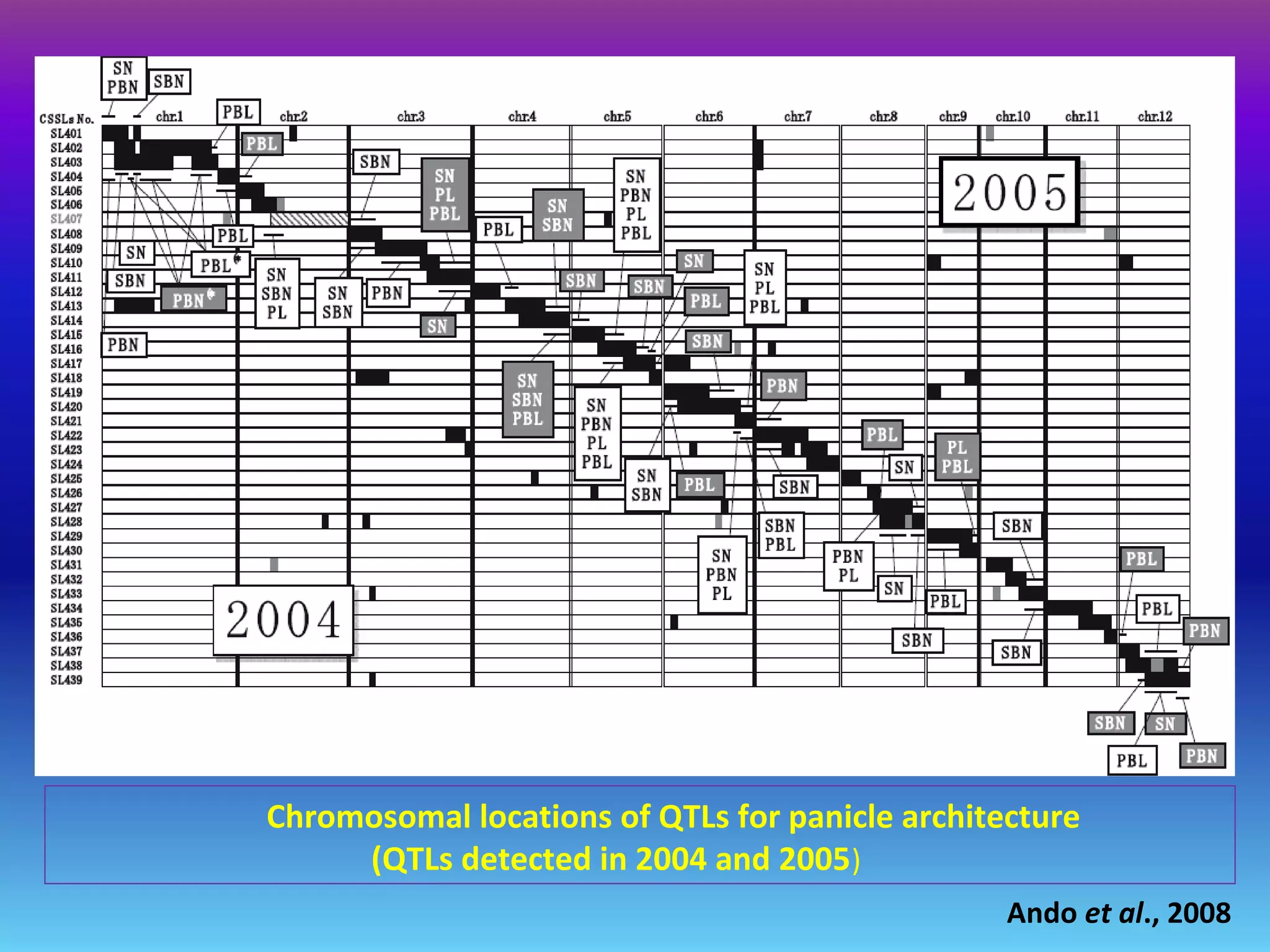
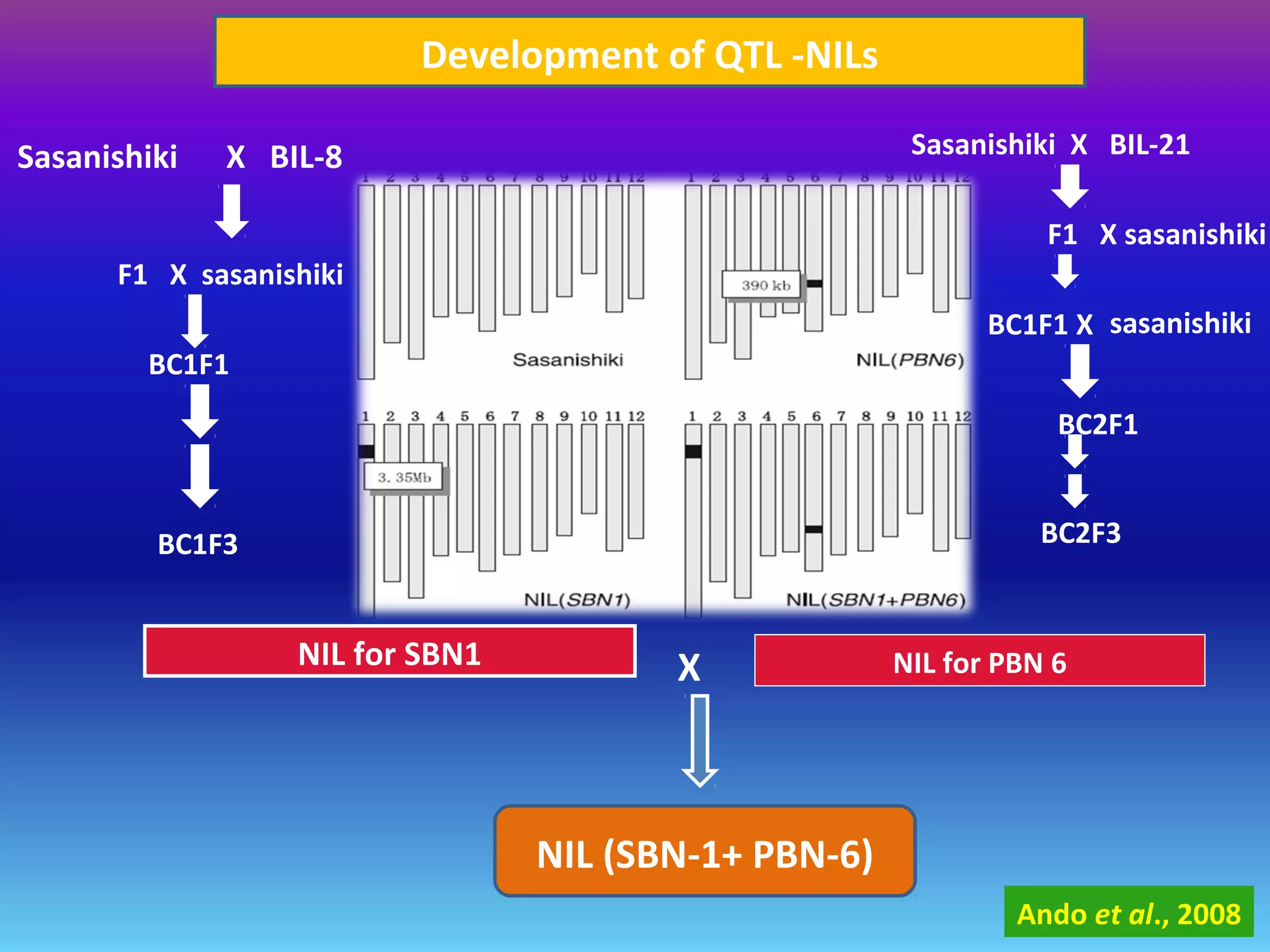
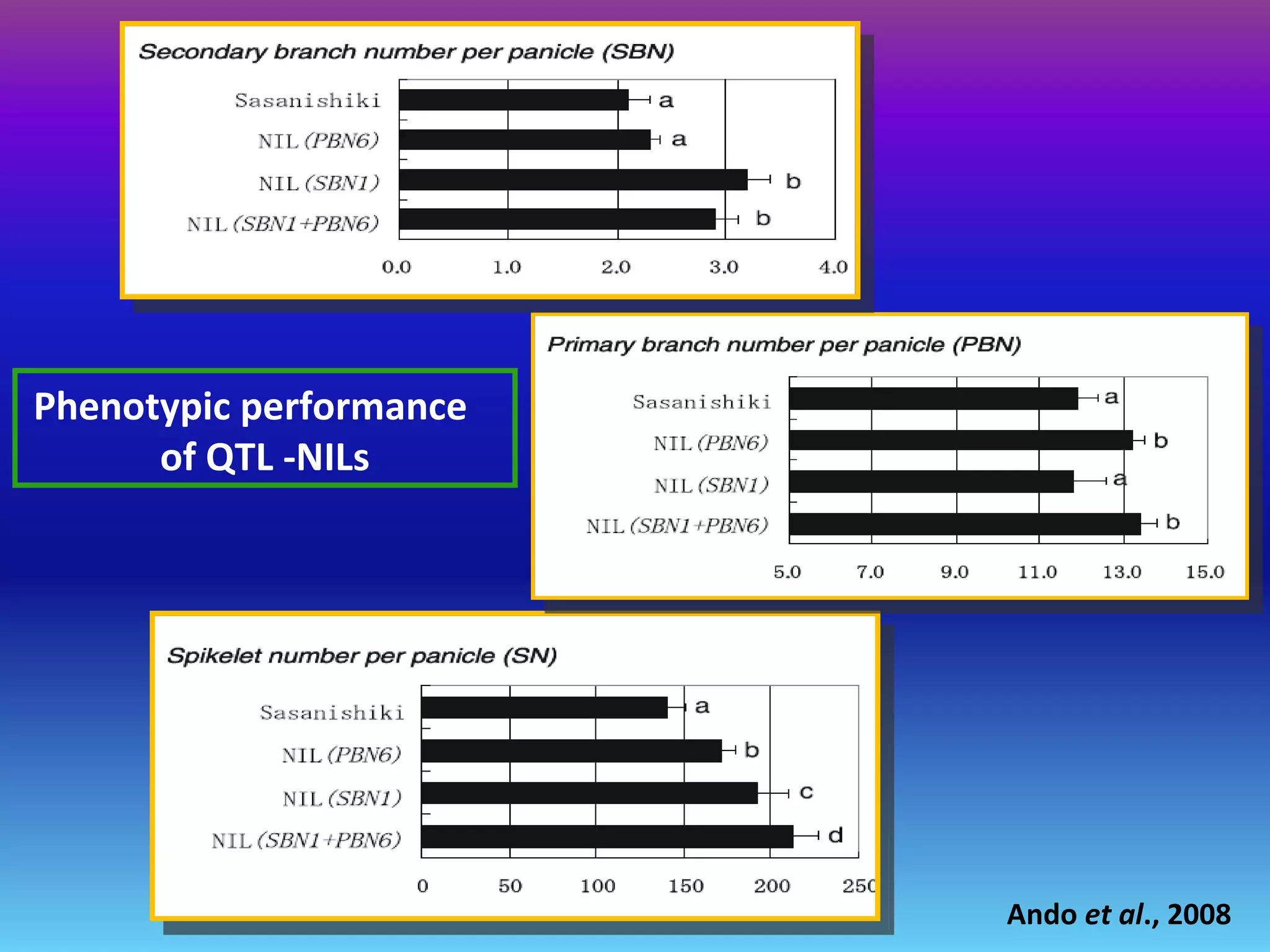
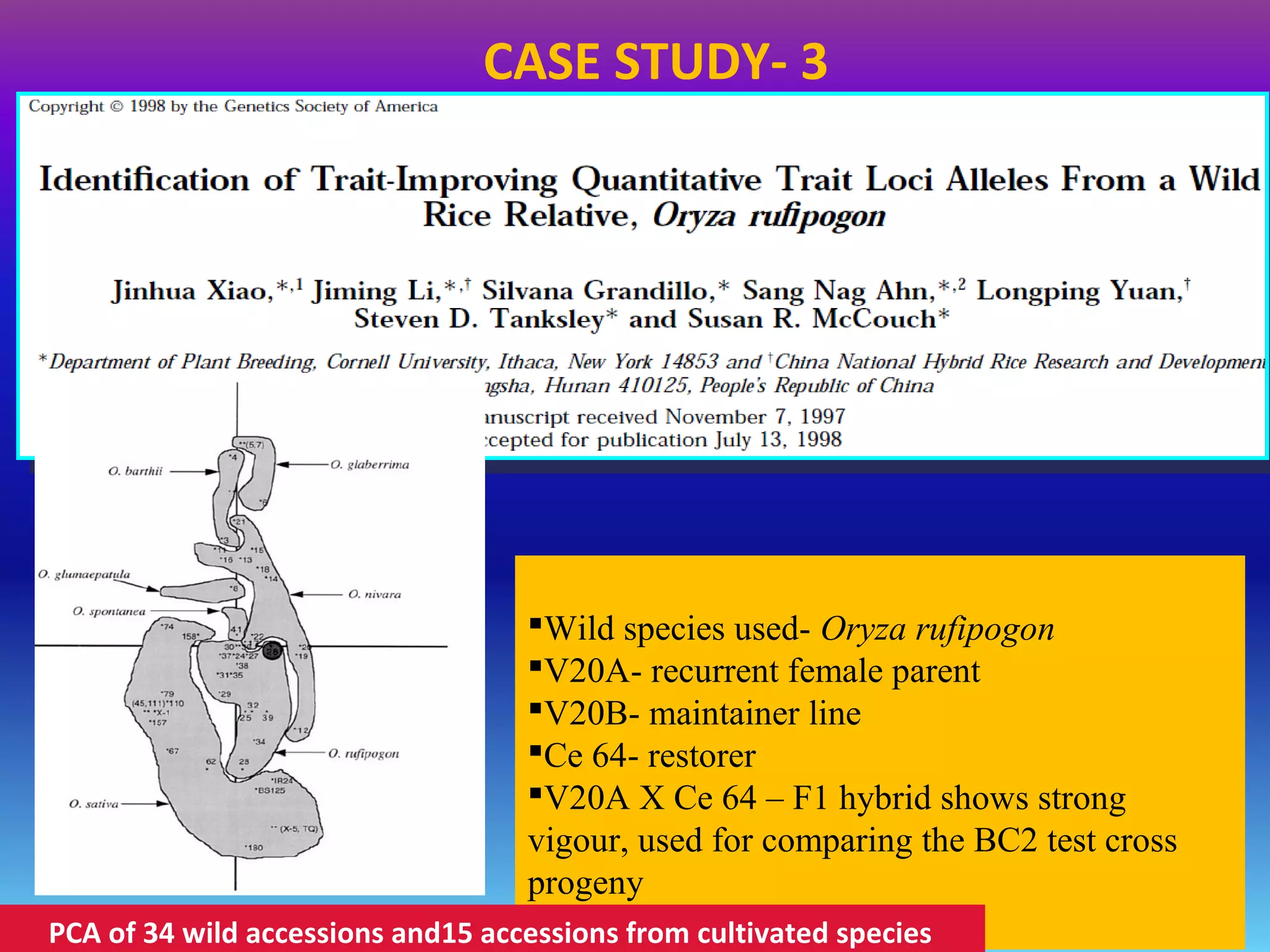
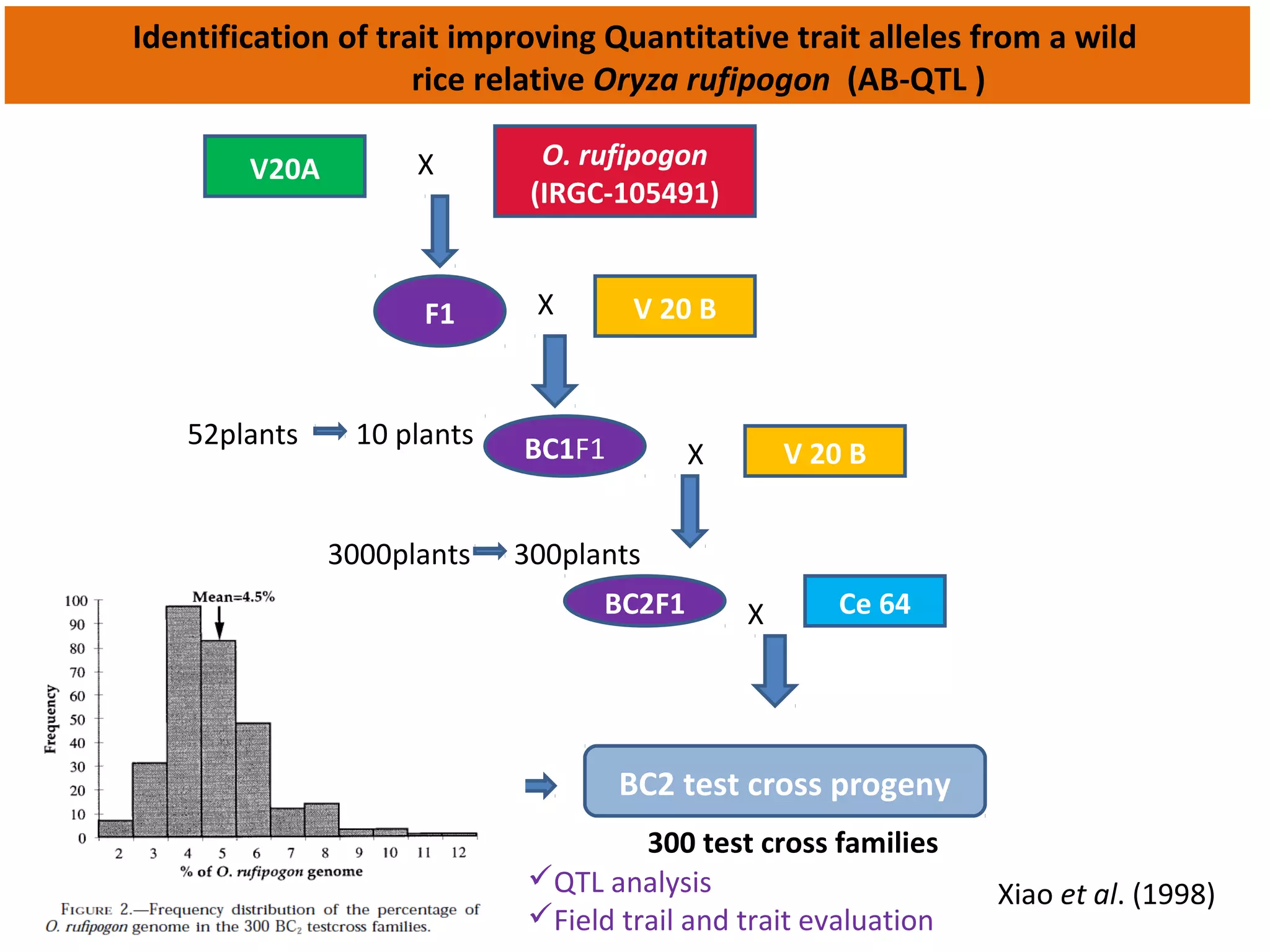
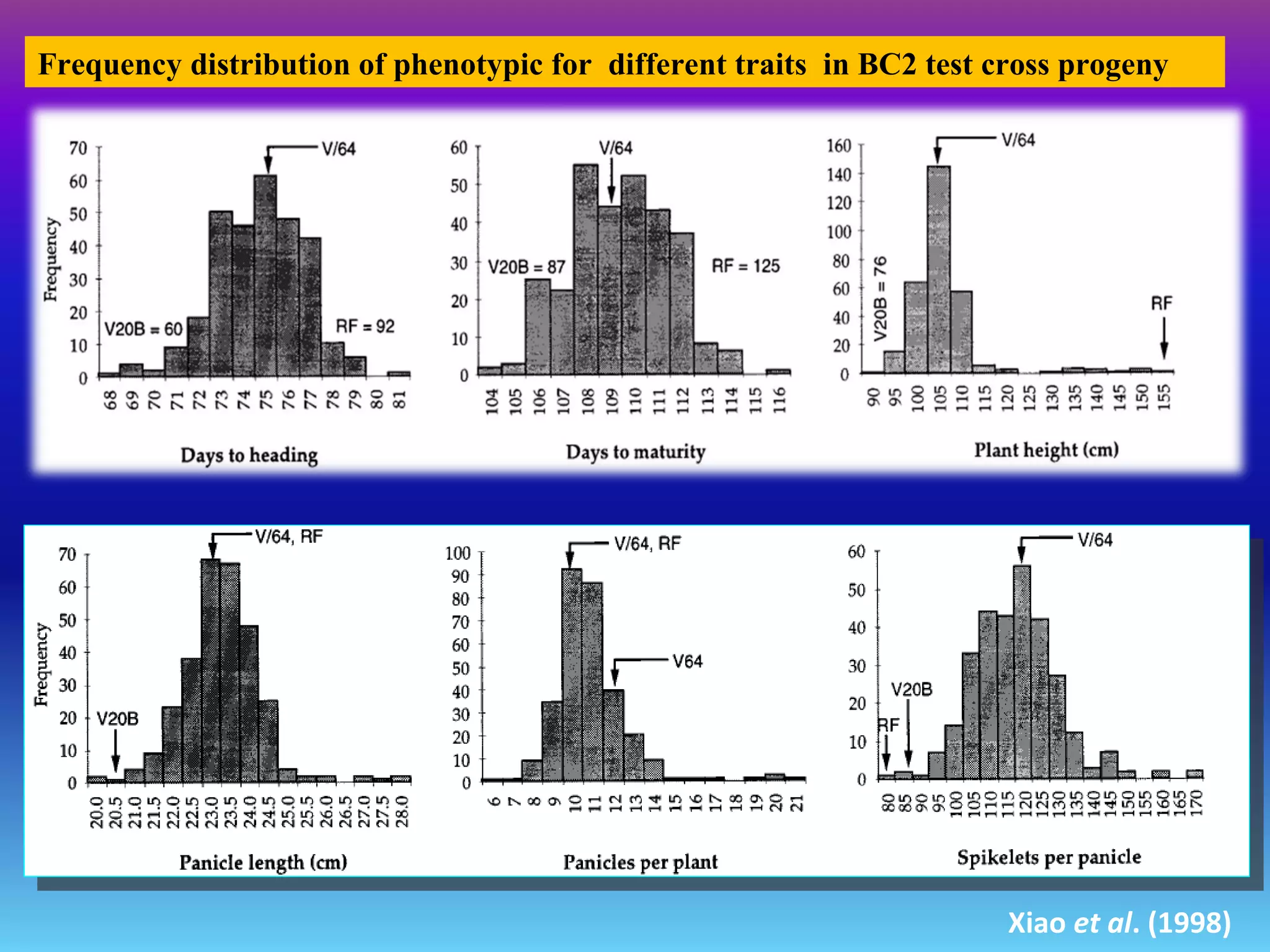
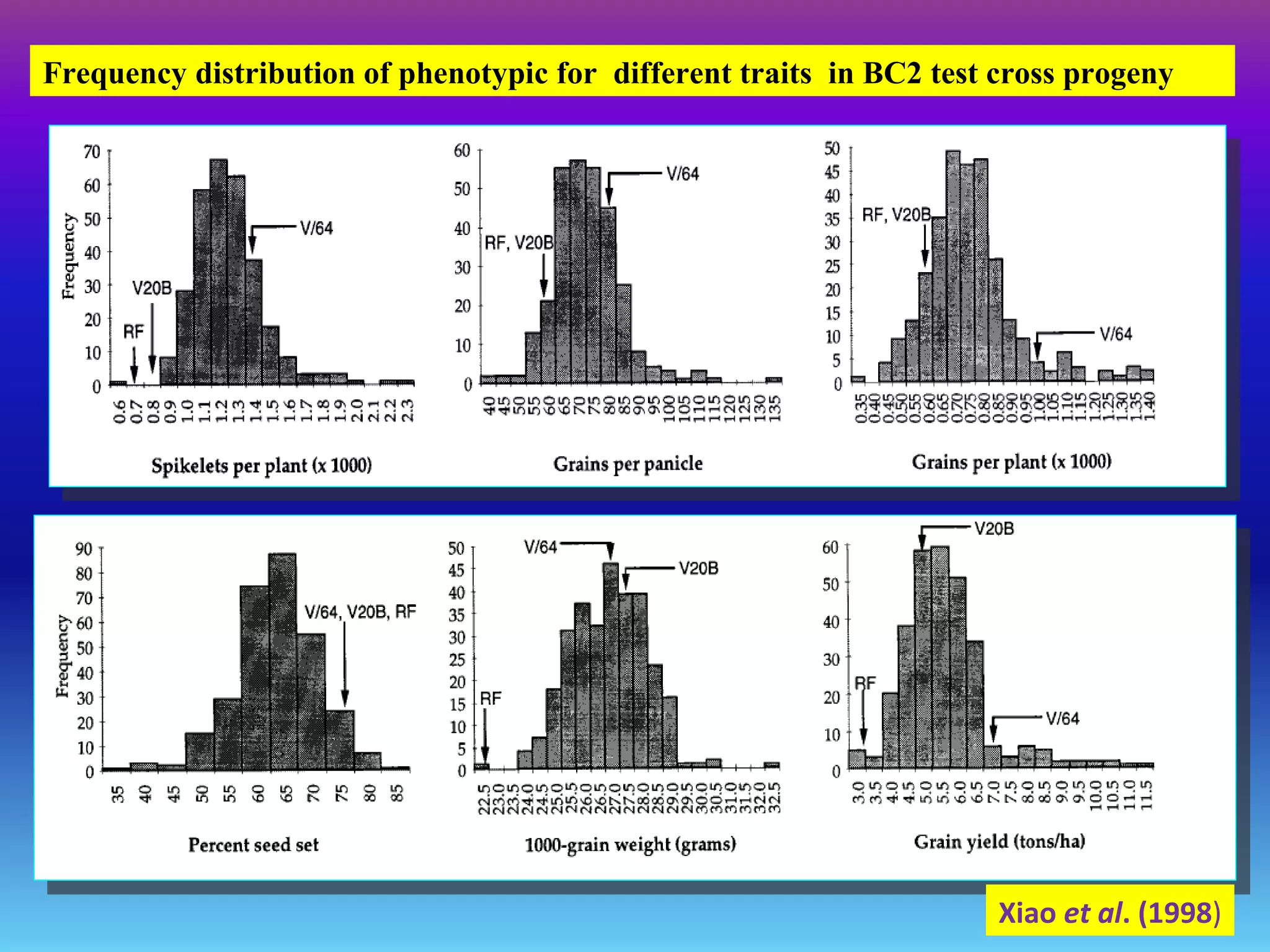
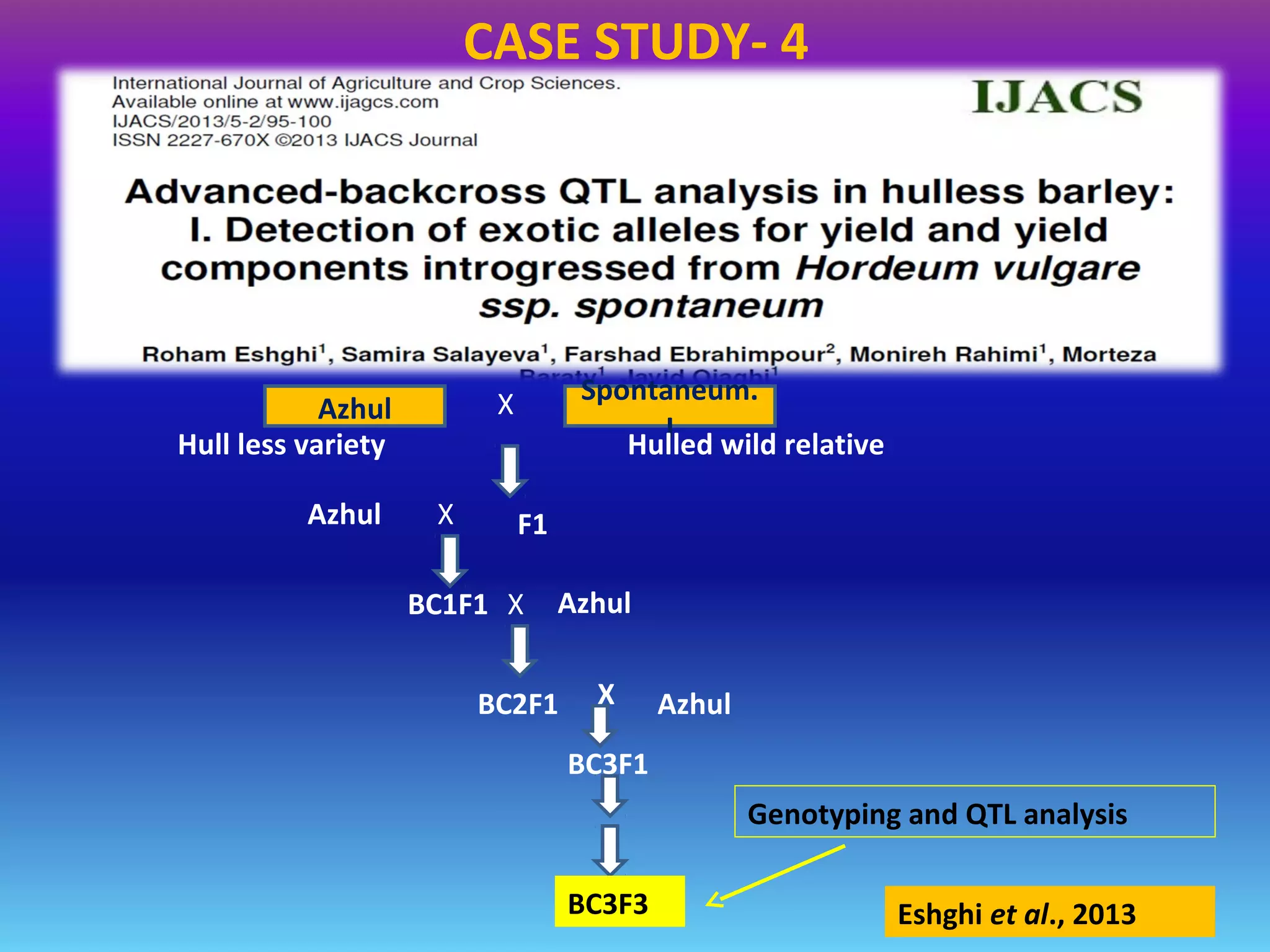
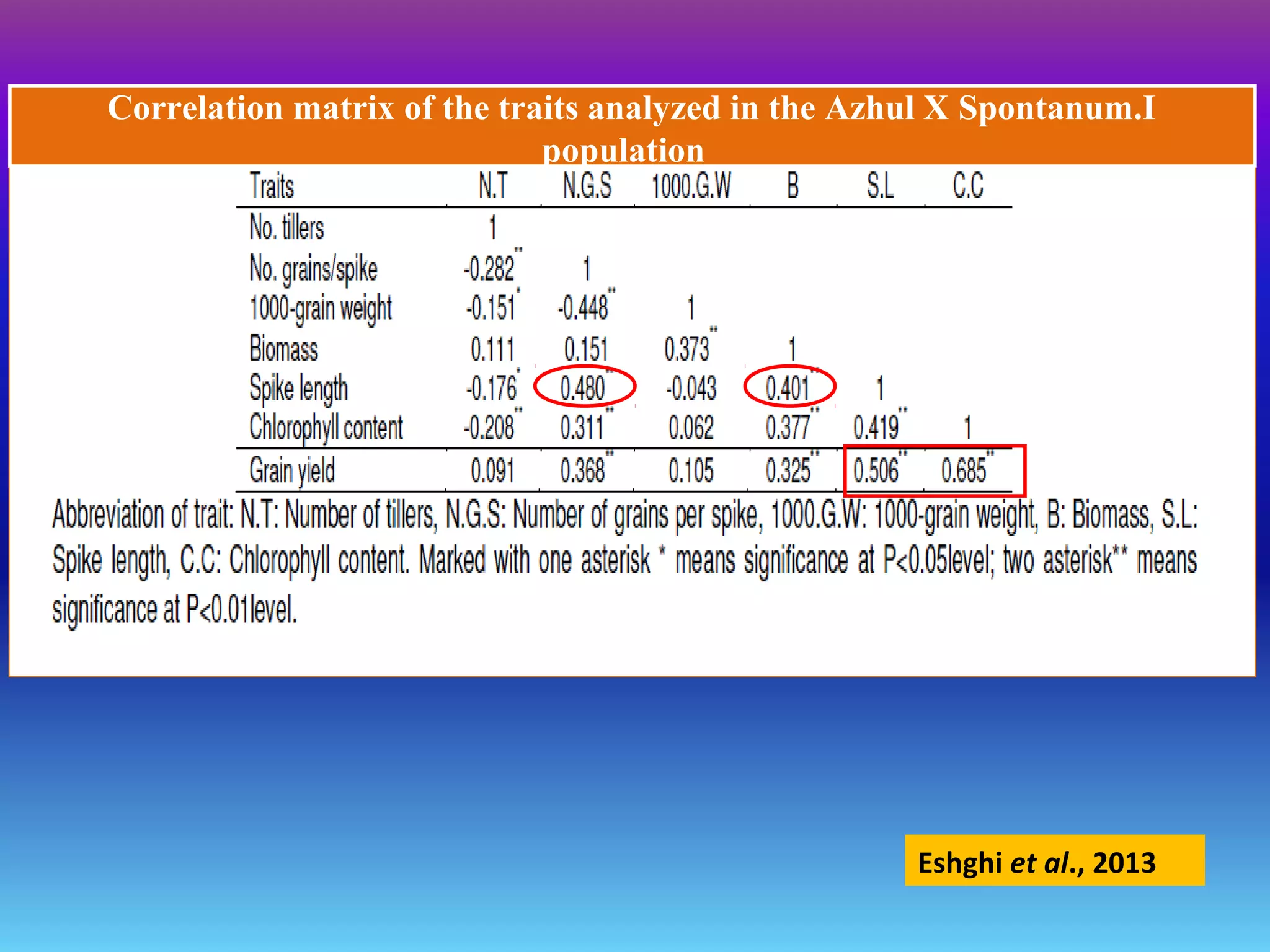
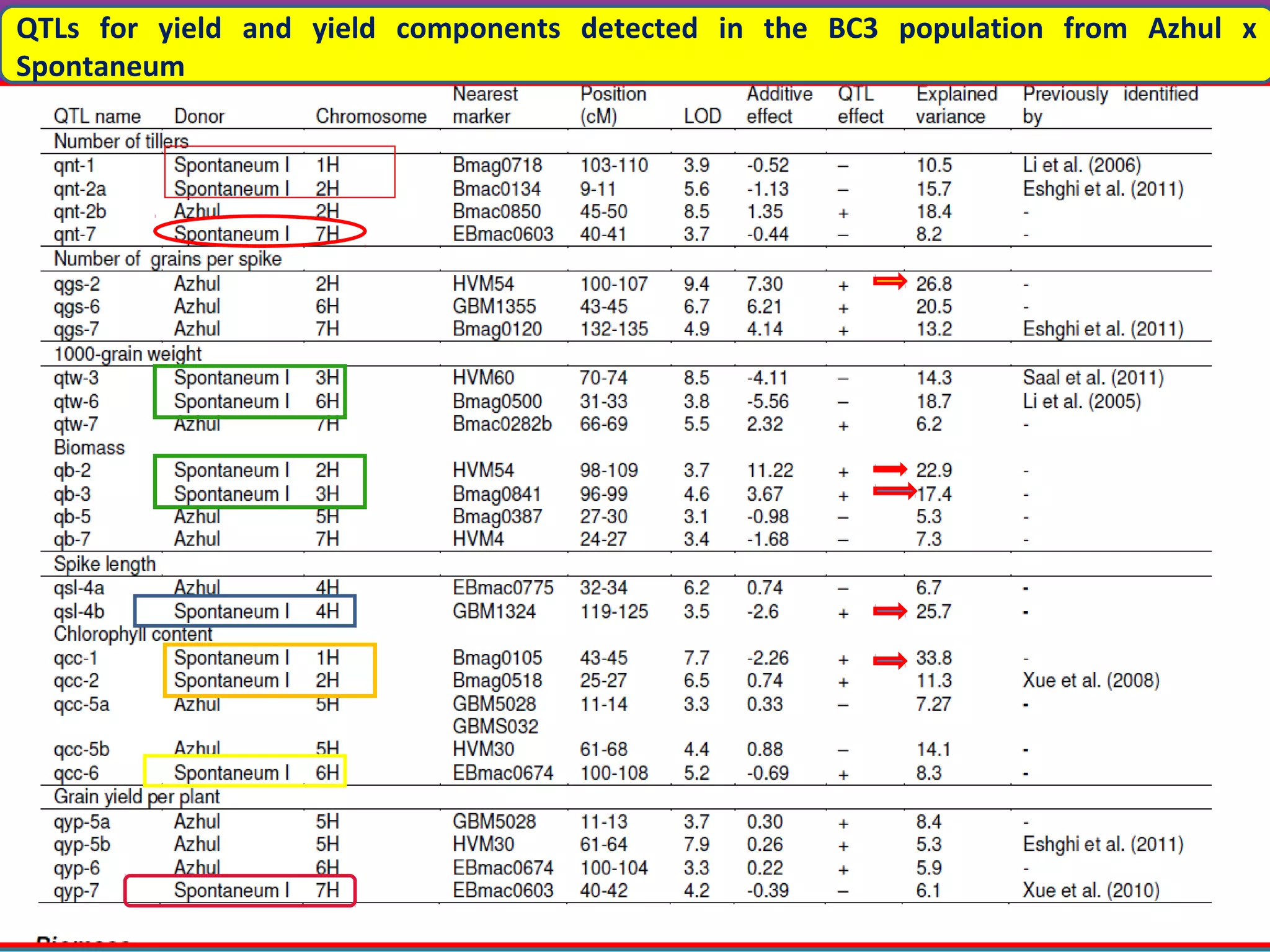
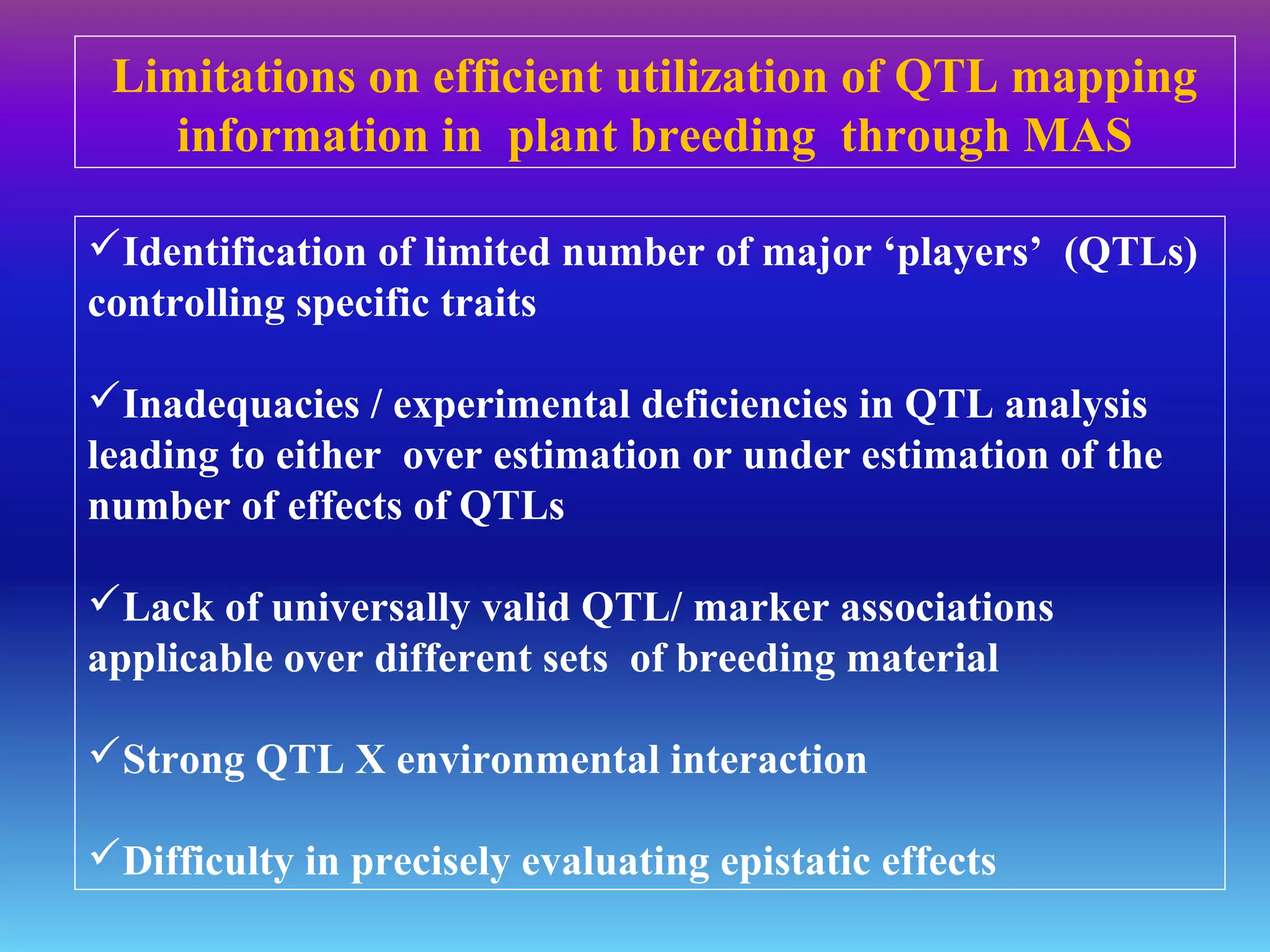
Marker-assisted selection (MAS) uses DNA markers linked to traits of interest to assist plant breeders in selecting desirable plants. MAS has advantages over phenotypic selection like enabling selection at early stages. MAS breeding schemes include marker-assisted backcrossing to introgress traits while minimizing linkage drag, and pyramiding to combine multiple genes/QTLs. Case studies demonstrate using MAS to develop rice varieties with submergence tolerance and improve yield traits. However, limitations include inconsistent QTL-marker associations across environments and difficulties evaluating complex trait genetics like epistasis. Future work aims to optimize MAS efficiency and integration with plant breeding.