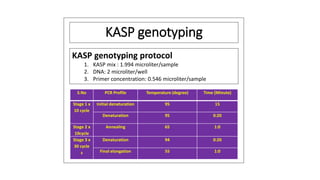
This document summarizes Shivendra Kumar's class presentation on SNP genotyping using KASP. It introduces SNP genotyping and the KASP platform. It describes using KASP to genotype a wheat mapping population derived from a cross between an introgression line containing stripe rust resistance genes and a susceptible cultivar. KASP markers were developed and used to map the resistance genes. One candidate resistance gene was identified and further analyzed through expression studies and development of a linked KASP marker. Recombinants were identified and confirmed through additional KASP genotyping.