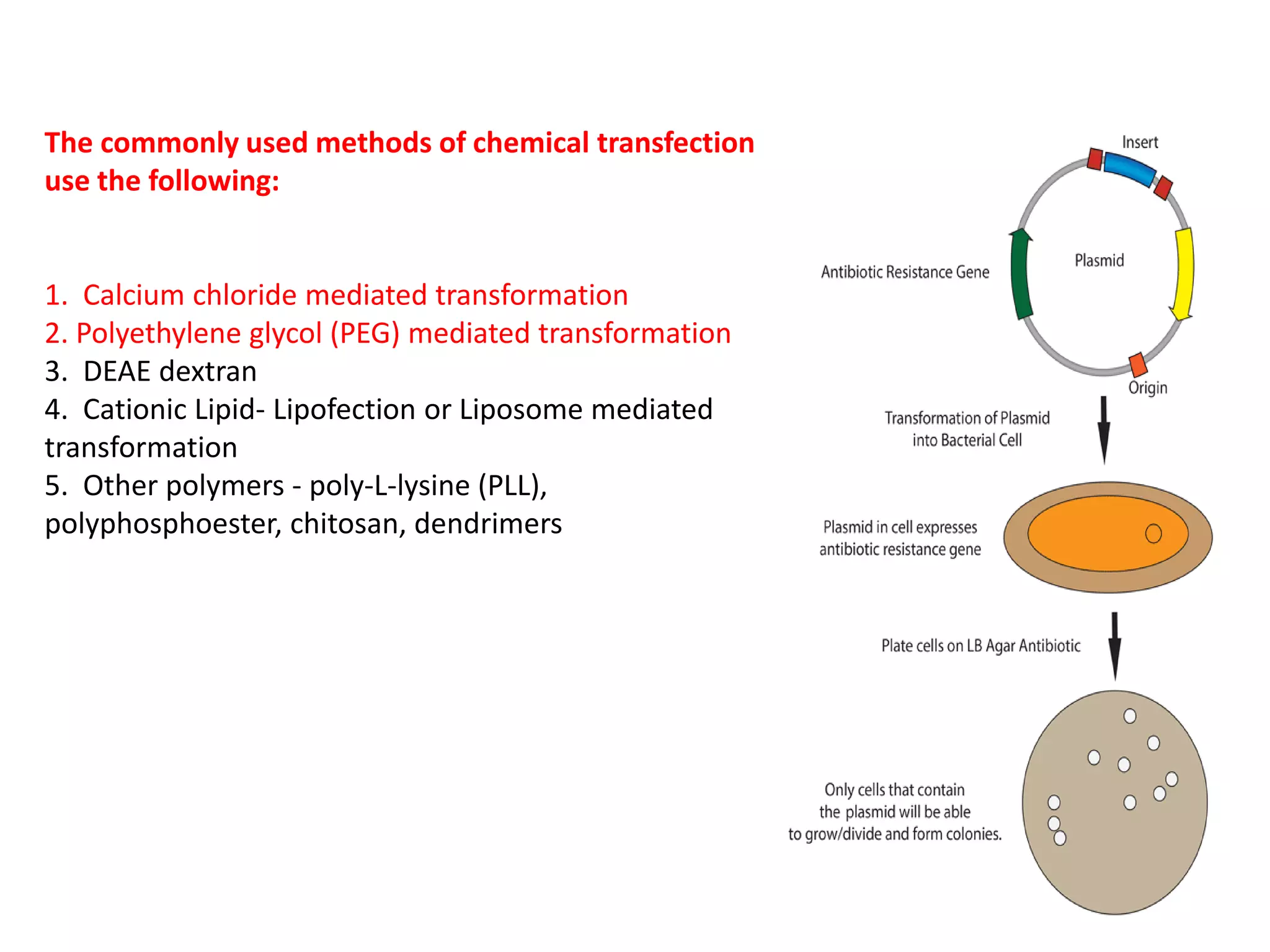
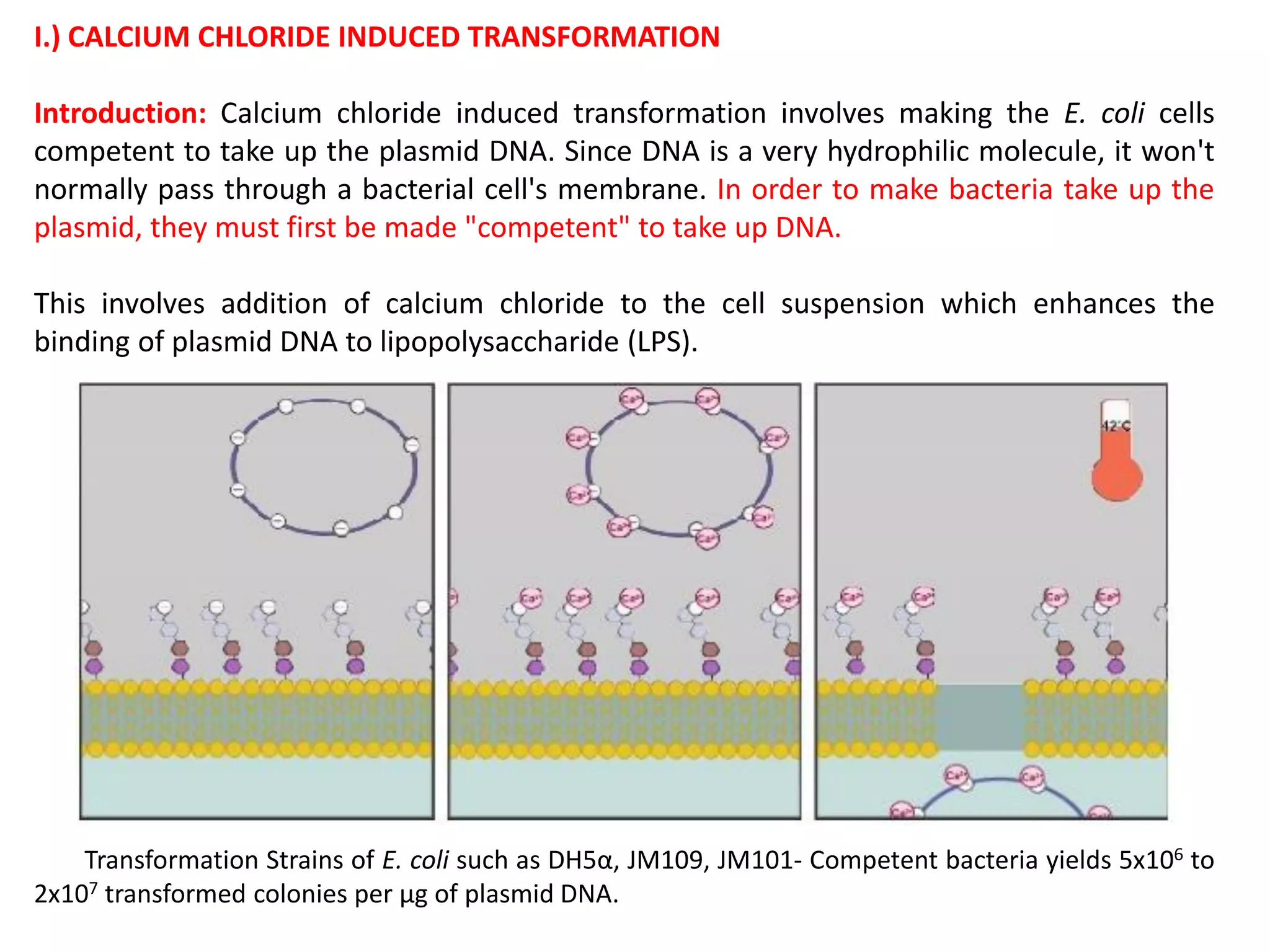
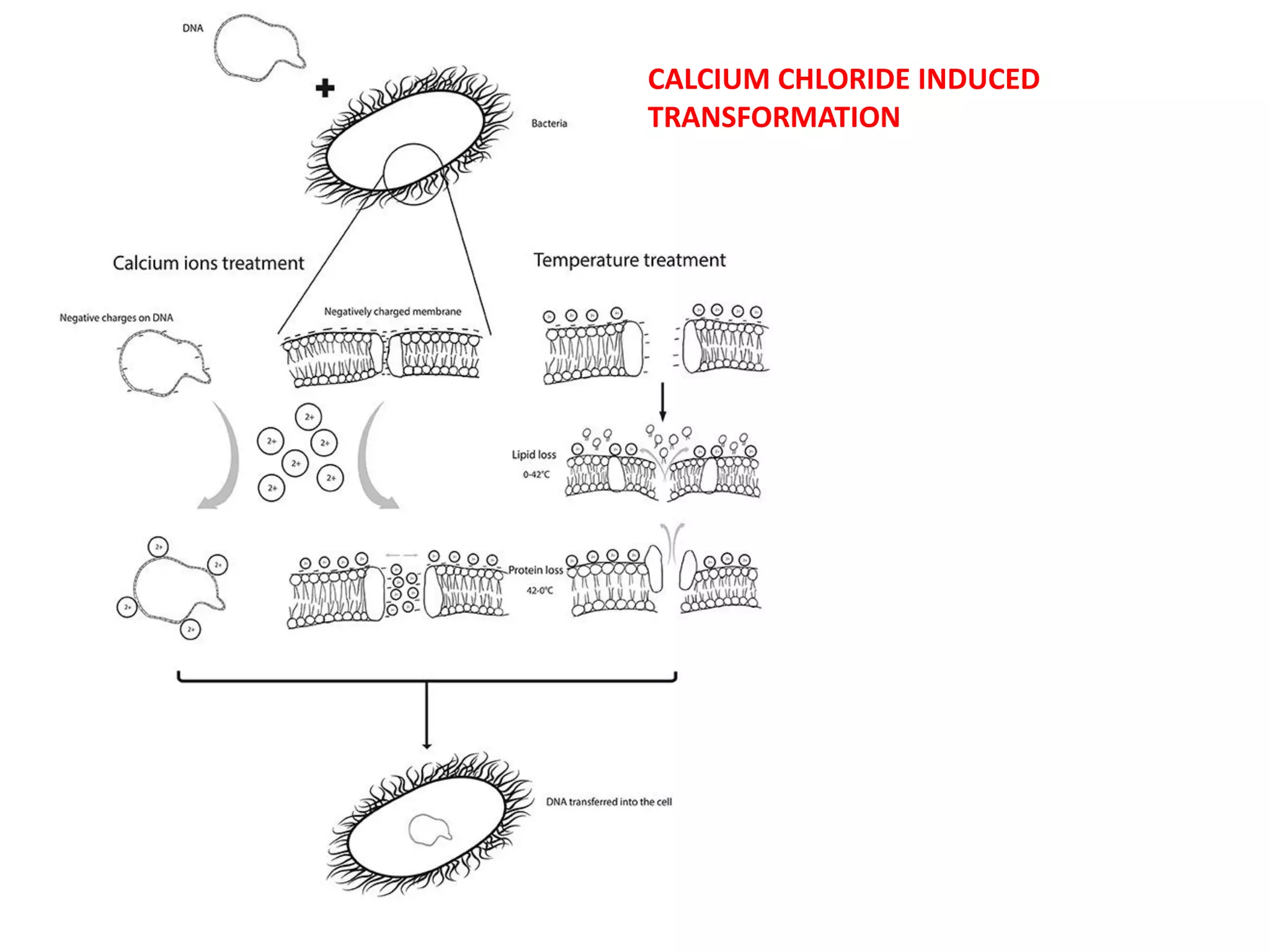
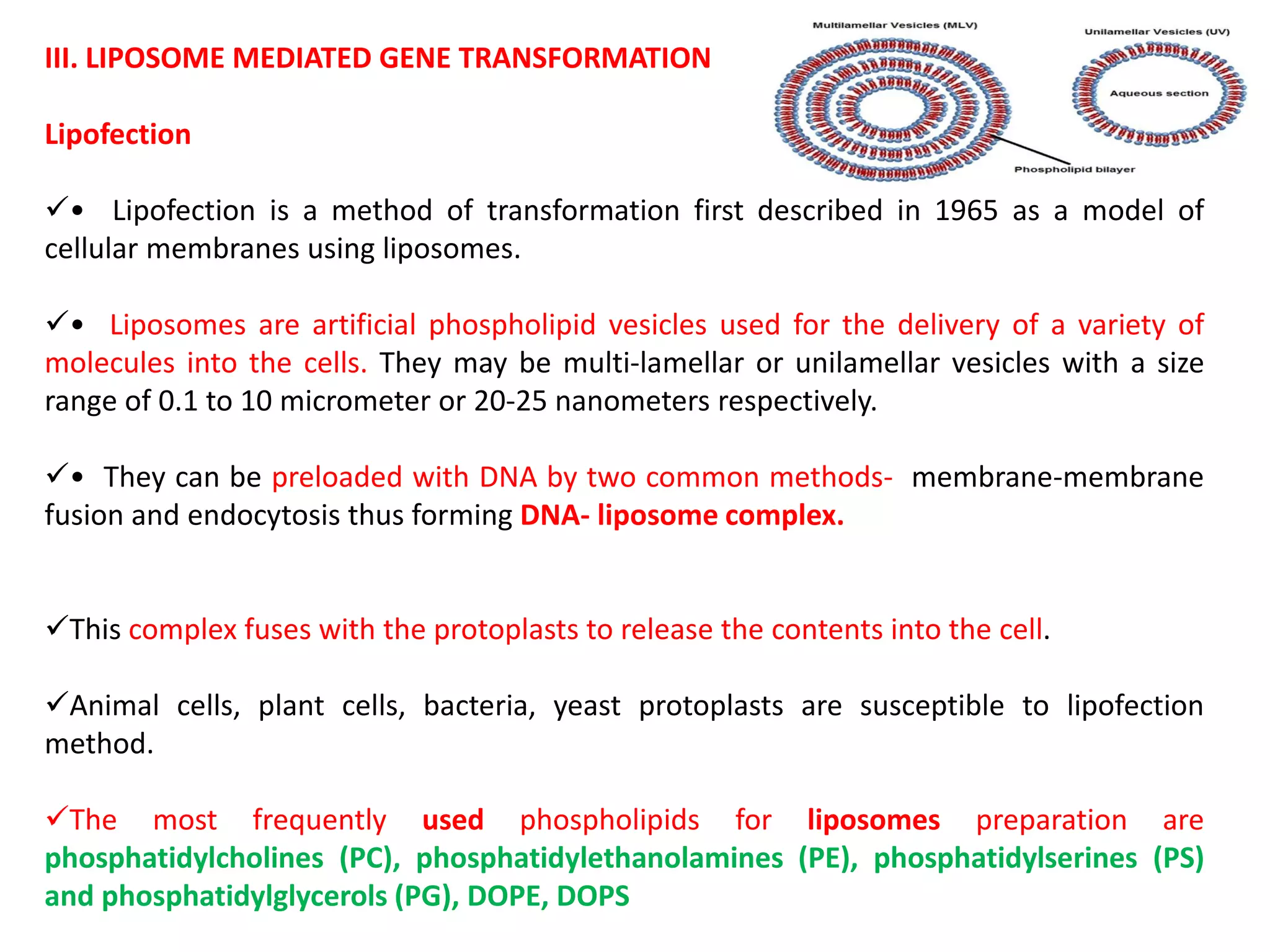
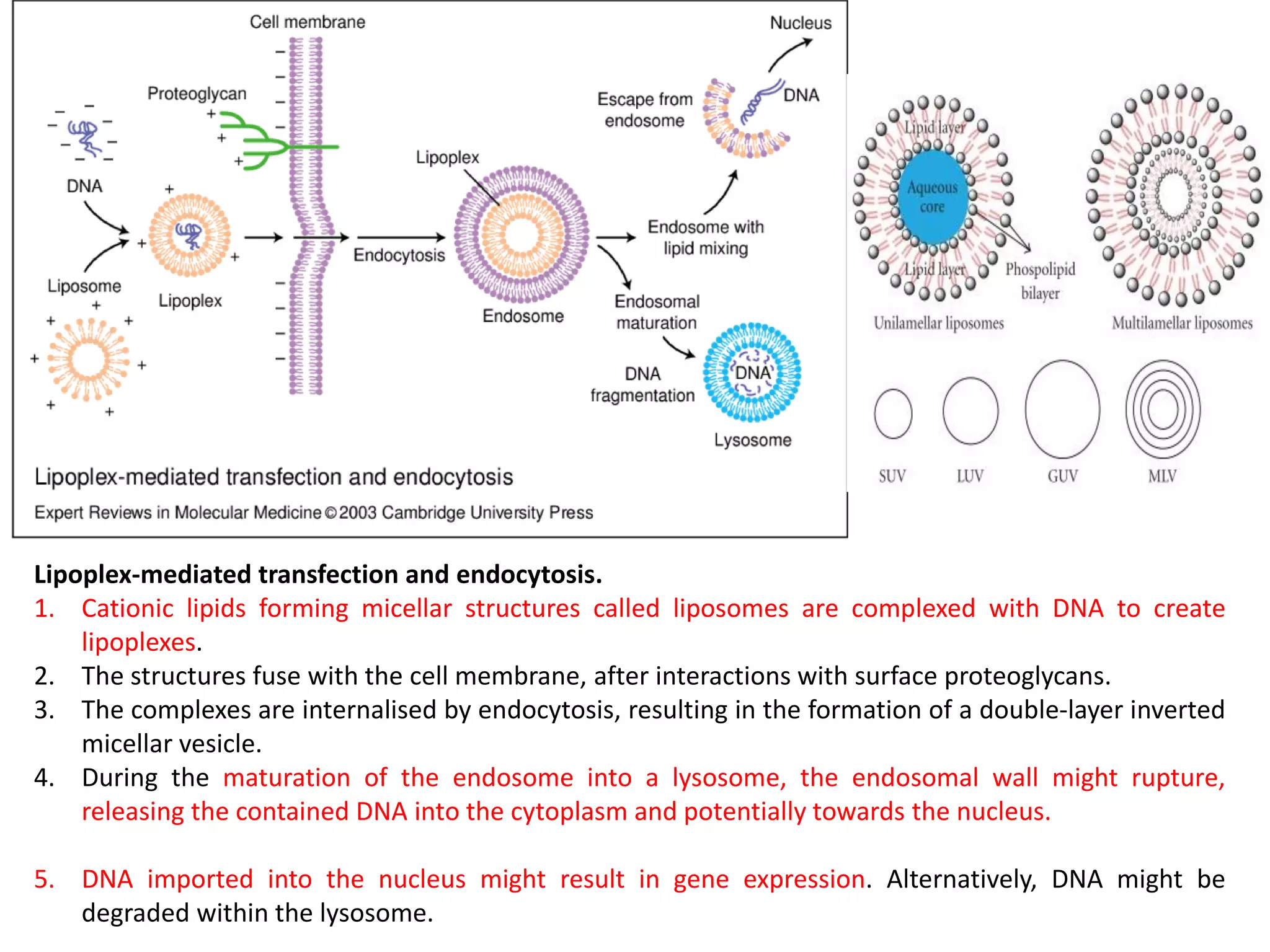
The document discusses various methods for introducing foreign DNA into host cells, including chemical, physical, and biological methods. It elaborates on the chemical transformation methods such as calcium chloride-mediated transformation and polyethylene glycol (PEG) method, detailing their principles, procedures, and advantages. Additionally, the document covers liposome-mediated gene transformation, highlighting the mechanisms of lipofection and the classification of liposomes based on their charge.