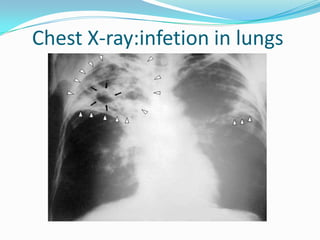
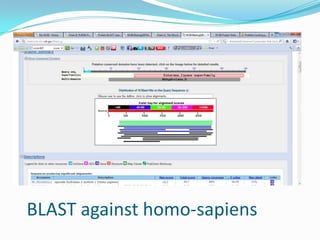
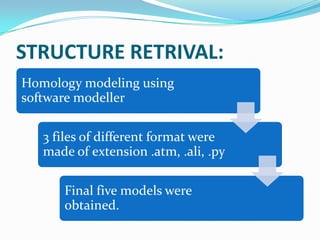
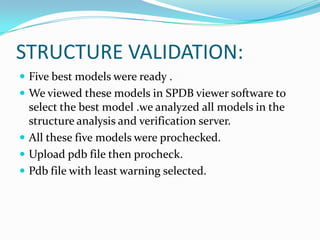
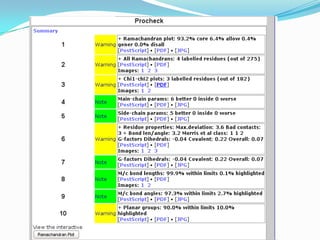
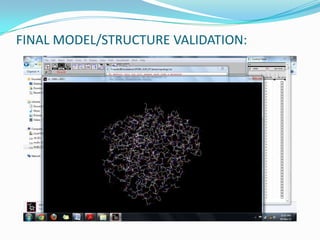
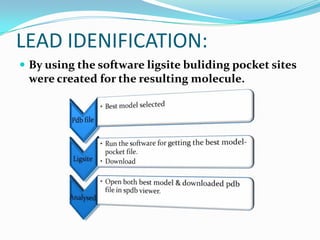
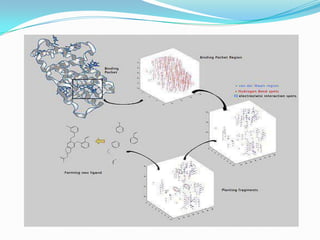
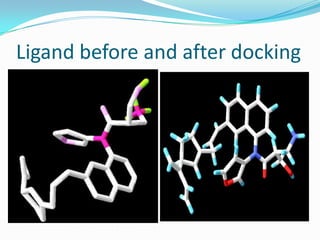
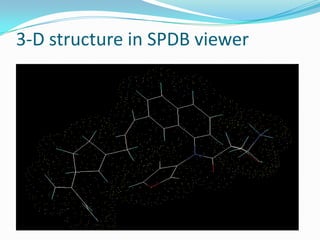
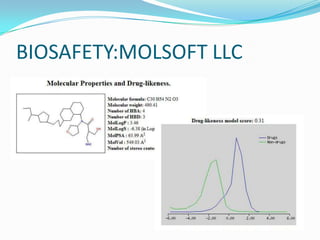
The document outlines the process of drug designing for tuberculosis, covering various aspects such as target identification, validation, structure retrieval, and final model development using computational methods. It details the steps involved in creating a prospective drug, including ligand binding affinity analysis and biosafety considerations. Additionally, it provides references to software and resources used throughout the drug design process.