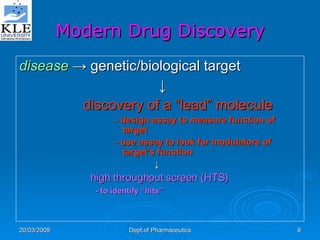
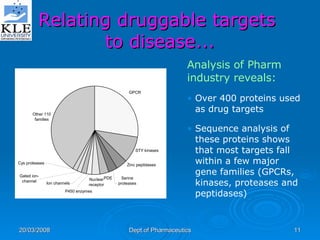
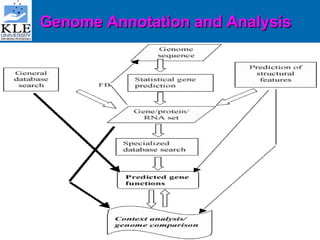
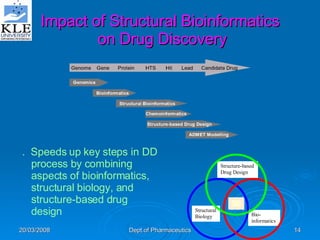
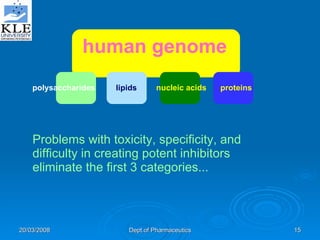
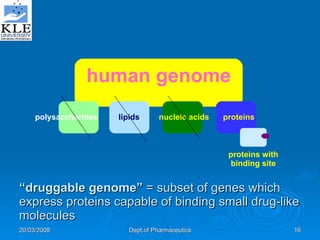
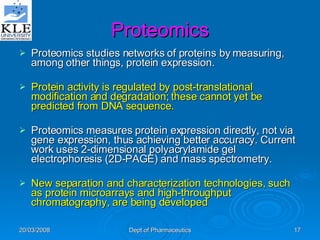
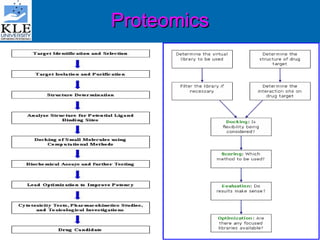
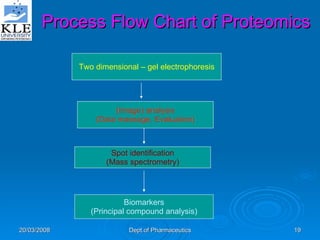
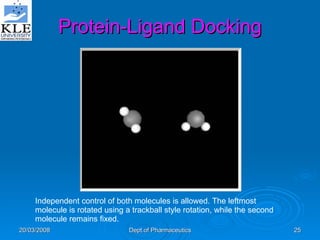
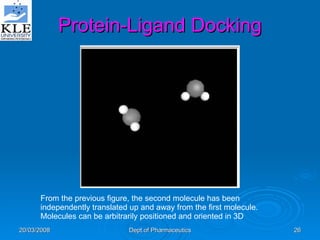
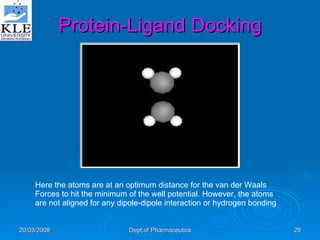
This document discusses genomics and proteomics based drug discovery. It explains that genomics involves sequencing genomes to understand gene functions and interactions, while proteomics studies protein expression and interactions. The document outlines how structural bioinformatics and techniques like protein-ligand docking can help in drug target identification and rational drug design. It also discusses how proteomics can aid in various stages of drug discovery like target identification and validation.