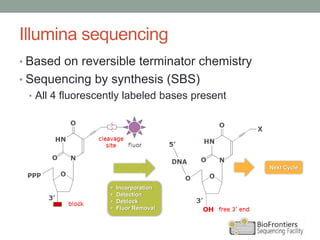
This document provides an overview of Illumina sequencing, including:
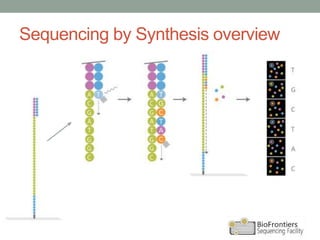
- Illumina sequencing uses a sequencing by synthesis (SBS) approach with reversible terminator chemistry. All four fluorescently labeled bases are present in each sequencing cycle.
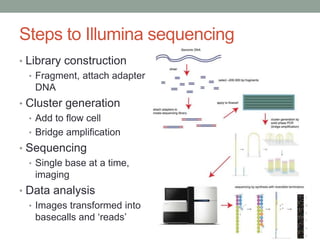
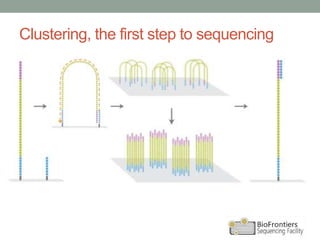
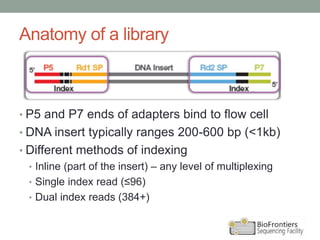
- Key steps include library construction, cluster generation, bridge amplification on the flow cell, and single-base sequencing imaging.
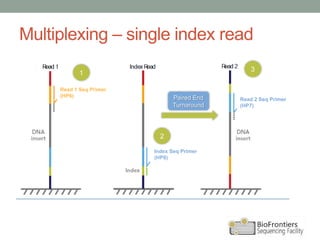
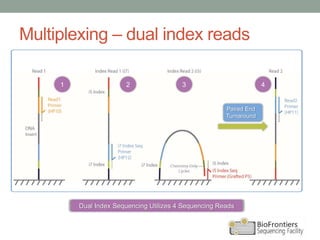
- Multiplexing allows indexing of multiple samples by attaching barcodes during library preparation. This enables pooled sequencing of many samples.
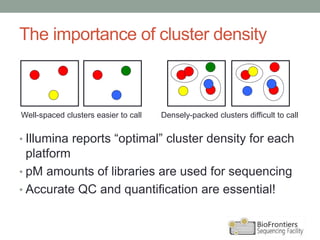
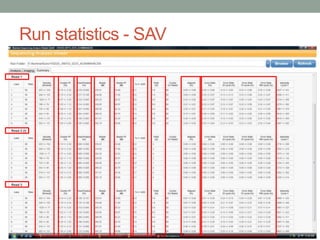
- Run statistics like number of reads, percentage of high-quality bases, and alignment rates provide information about run quality and performance.