1. The document discusses how genes work through the one-gene-one-enzyme hypothesis, which provides that each gene is responsible for producing a single enzyme.
2. It also covers the relationship between genes, proteins, and phenotypes. Mutations in a single nucleotide can alter protein function, as shown through studies of hemoglobin.
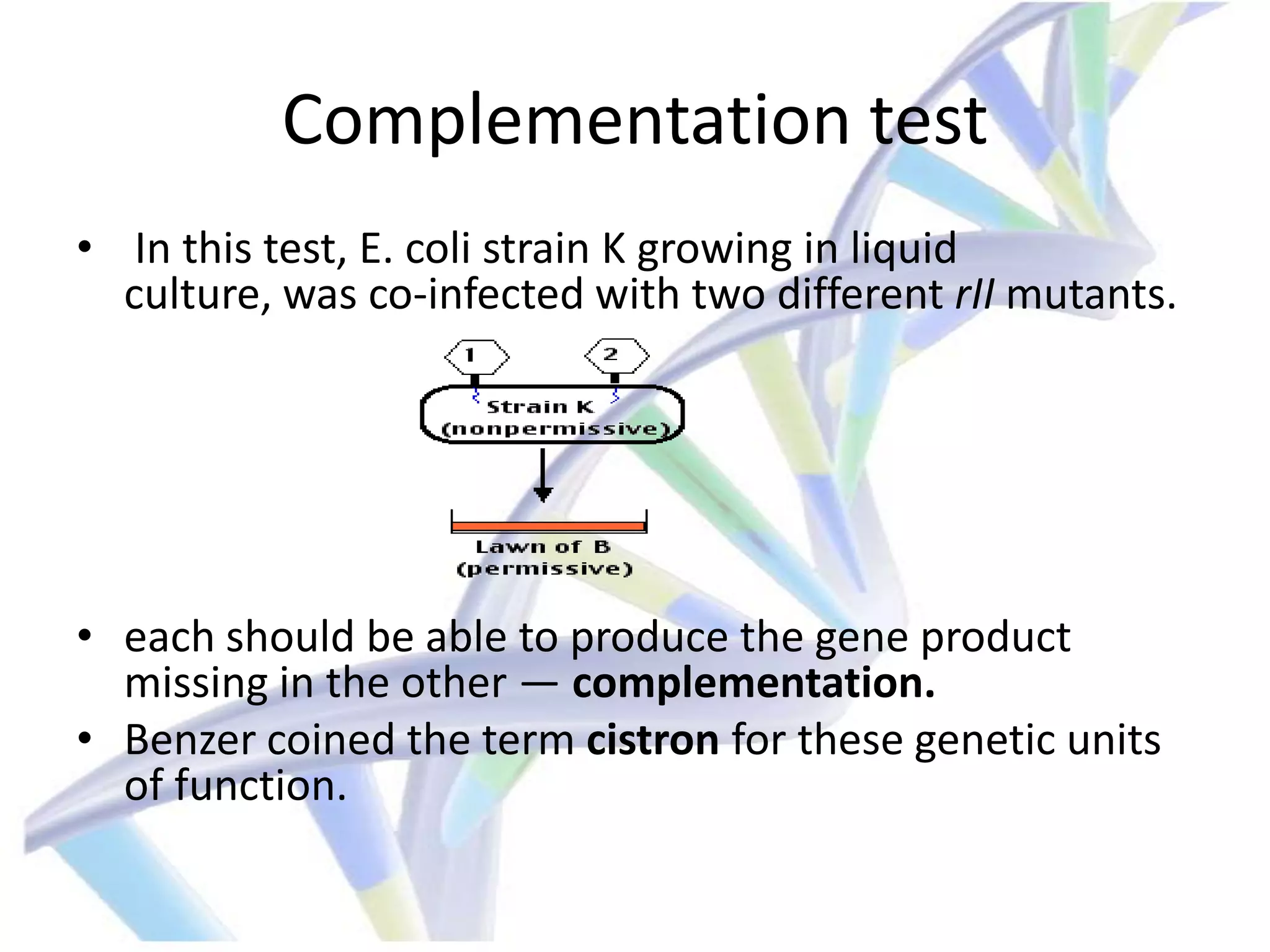
3. Experiments by Seymour Benzer using bacteriophage T4 helped disprove the "bead theory" by showing that the smallest units of mutation and recombination are single nucleotide pairs through analysis of point mutations and deletions within the rII gene.