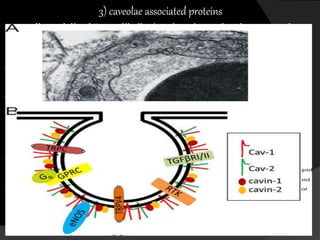
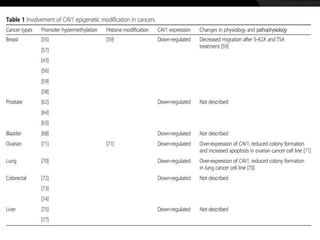
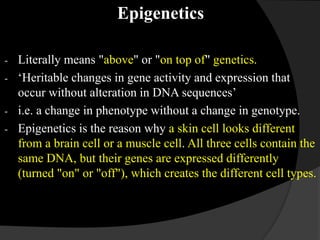
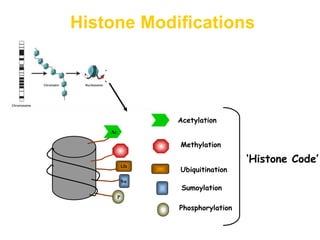
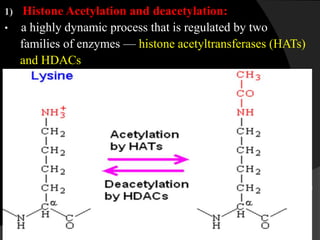
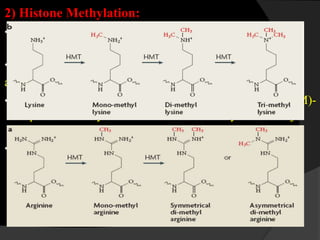
This document discusses epigenetic modifications of proteins. It begins by defining epigenetics and describing how epigenetic modifications can alter gene expression without changing DNA sequences. It then focuses on histone modifications like acetylation, methylation, ubiquitination, and phosphorylation. The document also examines epigenetic modifications of specific proteins - Sox2, CCAAT/enhancer binding protein a (C/EBPa), and caveolae associated proteins. It provides details on studies that analyzed the methylation of promoters for these genes in cancer cell lines and patient samples.





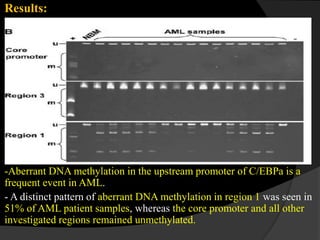
![Epigenetic Modification of C/EBPa
- One hundred forty-six bone marrow samples from AML patients.
- DNA and RNA Isolation.
- Assessment of C/EBPa methylation by COBRA.
[Combined Bisulfite Restriction Analysis (or COBRA) is a molecular
biology technique that allows for the sensitive quantification of DNA
methylation levels at a specific genomic locus on a DNA sequence in a
small sample of genomic DNA. The technique is a variation of bisulfite
sequencing, and combines bisulfite conversion based polymerase chain
reaction with restriction digestion. ]](https://image.slidesharecdn.com/epigeneticmodificationsofproteins-210410061945/85/Epigenetic-modifications-of-proteins-13-320.jpg)