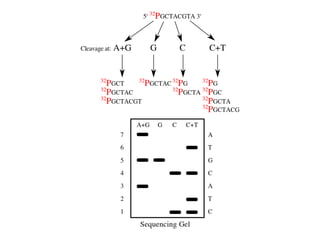
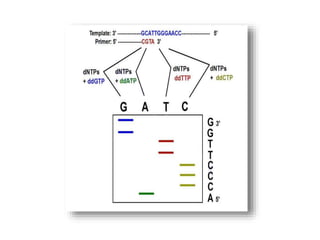
Maxam-Gilbert's sequencing method involves radiolabeling DNA at the 5' end and treating it with chemicals that cleave either adenine or guanine bases, or cytosine bases. This produces fragmented, radiolabeled DNA of varying lengths that can be separated by gel electrophoresis to determine the sequence. Sanger's method uses a single-stranded DNA template, a primer, DNA polymerase, normal dNTPs, and modified ddNTPs that terminate DNA strand elongation. Incorporation of a ddNTP causes DNA polymerase to stop, producing fragmented DNA of different lengths that reveal the sequence. Modern automated DNA sequencing builds on Sanger's chain termination method and uses fluorescence detection to determine sequences rapidly.