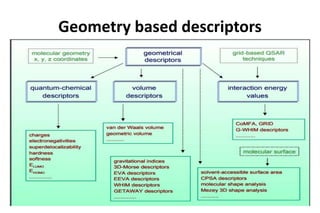
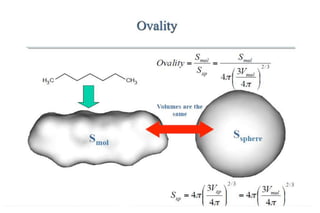
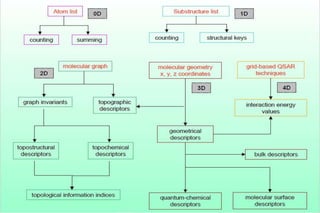
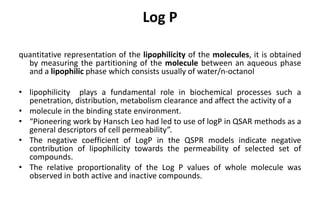
The document discusses different types of molecular descriptors that can be used to characterize chemical structures, including geometry-based descriptors, surface-based descriptors, structural fragments, and descriptors of lipophilicity. It provides examples of various quantum chemical descriptors, surface area descriptors, structural fragment descriptors, and ways to describe lipophilicity and log P values. The document also briefly mentions the varying computational requirements of different descriptor types.