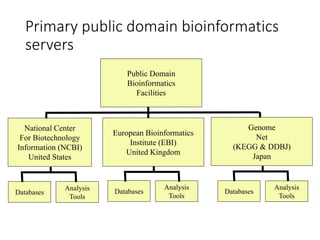
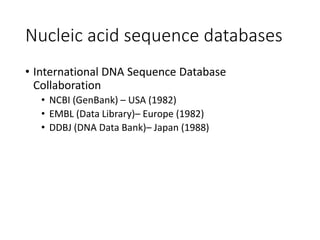
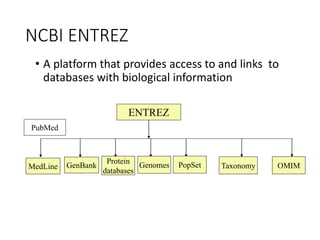
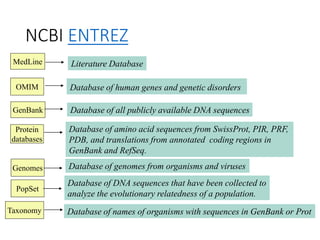
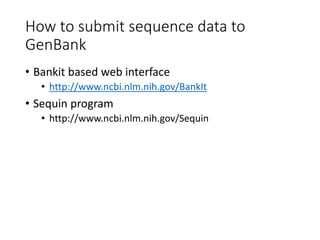
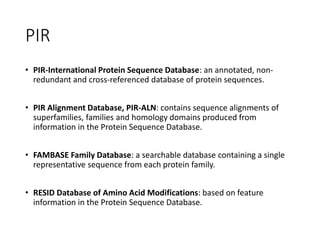
The document discusses several key biological databases and resources for archiving and retrieving genomic and protein information. It describes primary public databases located in Europe, the US, and Japan that house sequence data, analysis tools, and literature. Key databases mentioned include GenBank, EMBL, DDBJ for nucleic acid sequences, Swiss-Prot and PIR for proteins, and PDB for protein structures. NCBI and its Entrez portal provide integrated access to these and additional databases like PubMed, OMIM, and Taxonomy. The document outlines how to submit data to GenBank and search various protein and literature databases.