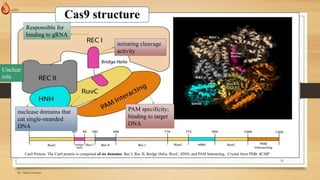
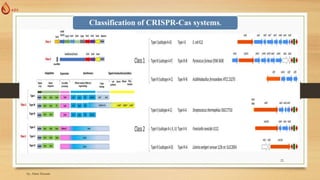
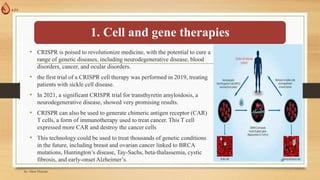
The document discusses the introduction, history, and applications of CRISPR technology, which is a powerful genetic editing tool derived from bacterial immune systems. Key milestones include its discovery by Francisco Mojica, the development of CRISPR-Cas9 for gene editing, and its utilization in various fields such as medicine, agriculture, and diagnostics. The potential of CRISPR technology to revolutionize treatments for genetic diseases and contribute to advancements in crop resilience and bioenergy is highlighted.