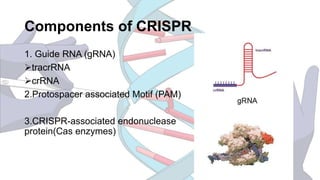
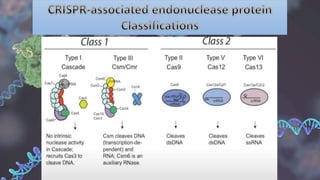
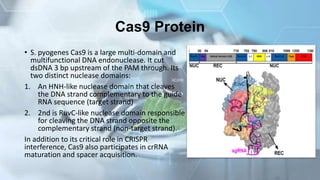
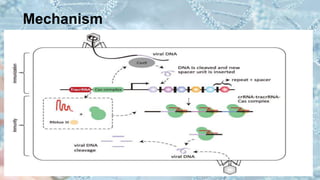
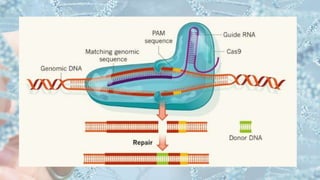
CRISPR is a groundbreaking genome editing tool derived from bacterial immune systems, enabling precise modifications of DNA through components like guide RNA and Cas9 proteins. Its development has evolved since its initial discovery in 1987, culminating in practical applications for gene editing, disease prevention, and agricultural improvements. While it offers significant advantages such as precision and versatility, ethical concerns and potential off-target effects present challenges to its widespread adoption.