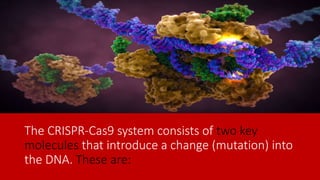
CRISPR-Cas9 is an advanced genome editing tool that allows for precise modifications of DNA, using a combination of the Cas9 enzyme and guide RNA (gRNA) to target and edit specific genomic sequences. The technology originated from bacteria's immune response to viruses and has been adapted for use in various organisms, including humans. While CRISPR-Cas9 presents significant potential for research and therapy, ethical and legal considerations arise, particularly regarding germline modifications.