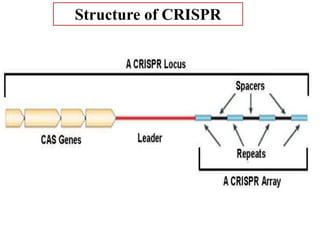
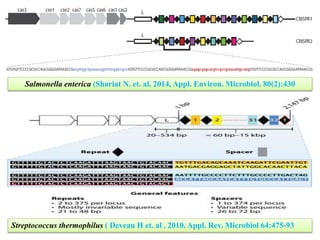
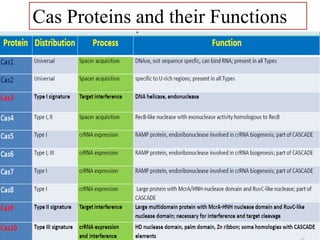
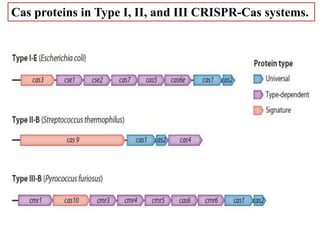
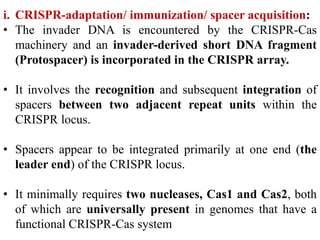
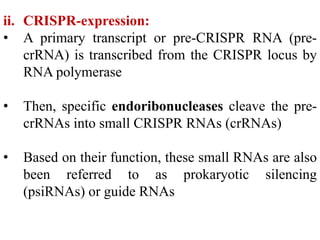
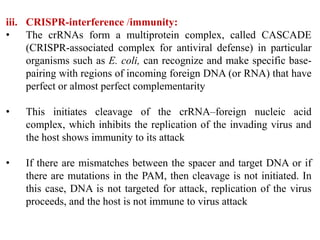
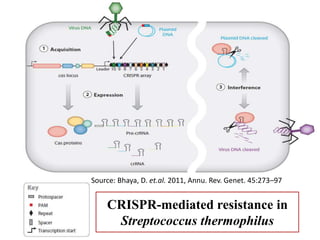
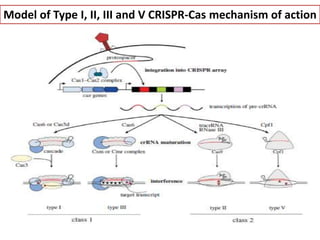
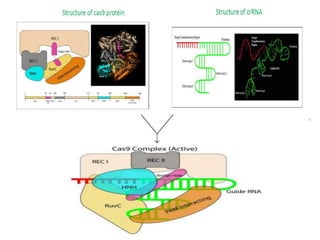
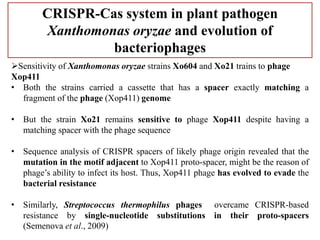
CRISPR-Cas systems provide bacteria with acquired immunity against viruses and plasmids. CRISPR loci contain repeating sequences separated by unique spacer sequences that are derived from invading genetic elements. Cas proteins process CRISPR RNA transcripts from the loci into small CRISPR RNAs that guide the degradation of invading nucleic acids based on sequence complementarity. This three-stage CRISPR-Cas immune response of adaptation, expression, and interference integrates new spacers and uses CRISPR RNAs to target matching foreign genetic elements. CRISPR-Cas systems are found in many bacteria and archaea and can be exploited for applications like bacterial typing, evolution studies, and generating phage resistance.