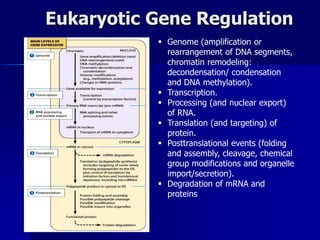
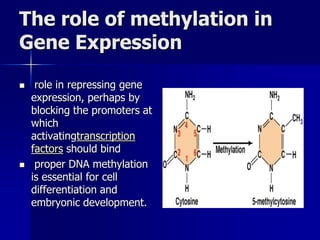
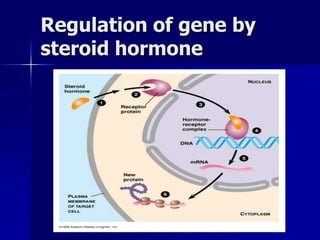
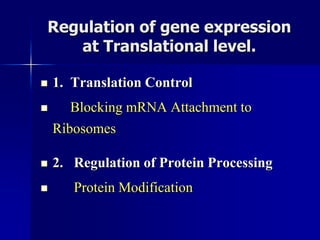
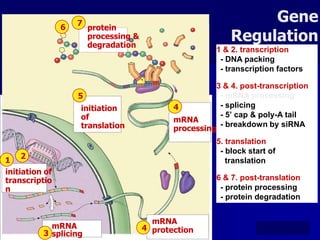
Regulation of gene expression in eukaryotes can occur at multiple levels:
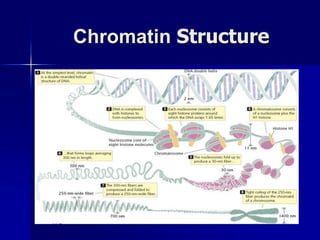
(1) The genome level through chromatin remodeling, DNA methylation, and gene amplification or rearrangement.
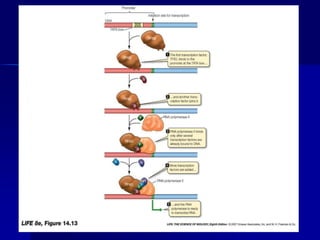
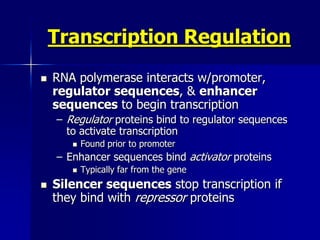
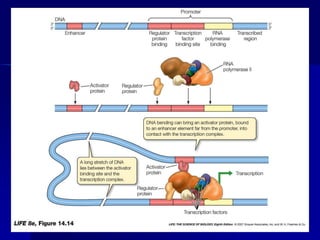
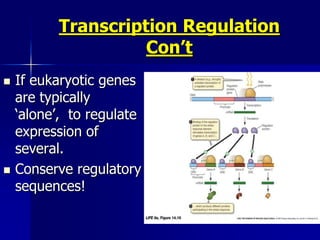
(2) The transcription level where transcription factors and cis-acting elements like promoters, enhancers, and silencers regulate RNA polymerase binding and activity.
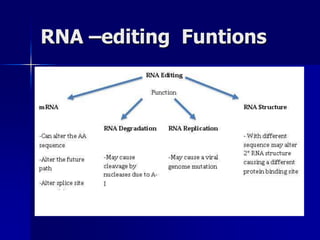
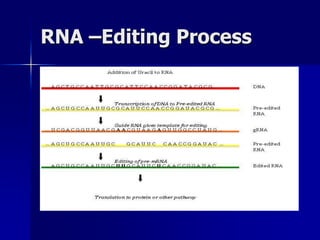
(3) Post-transcriptional processing of transcripts through RNA editing, alternative splicing, and transcript degradation which expands proteomic complexity.