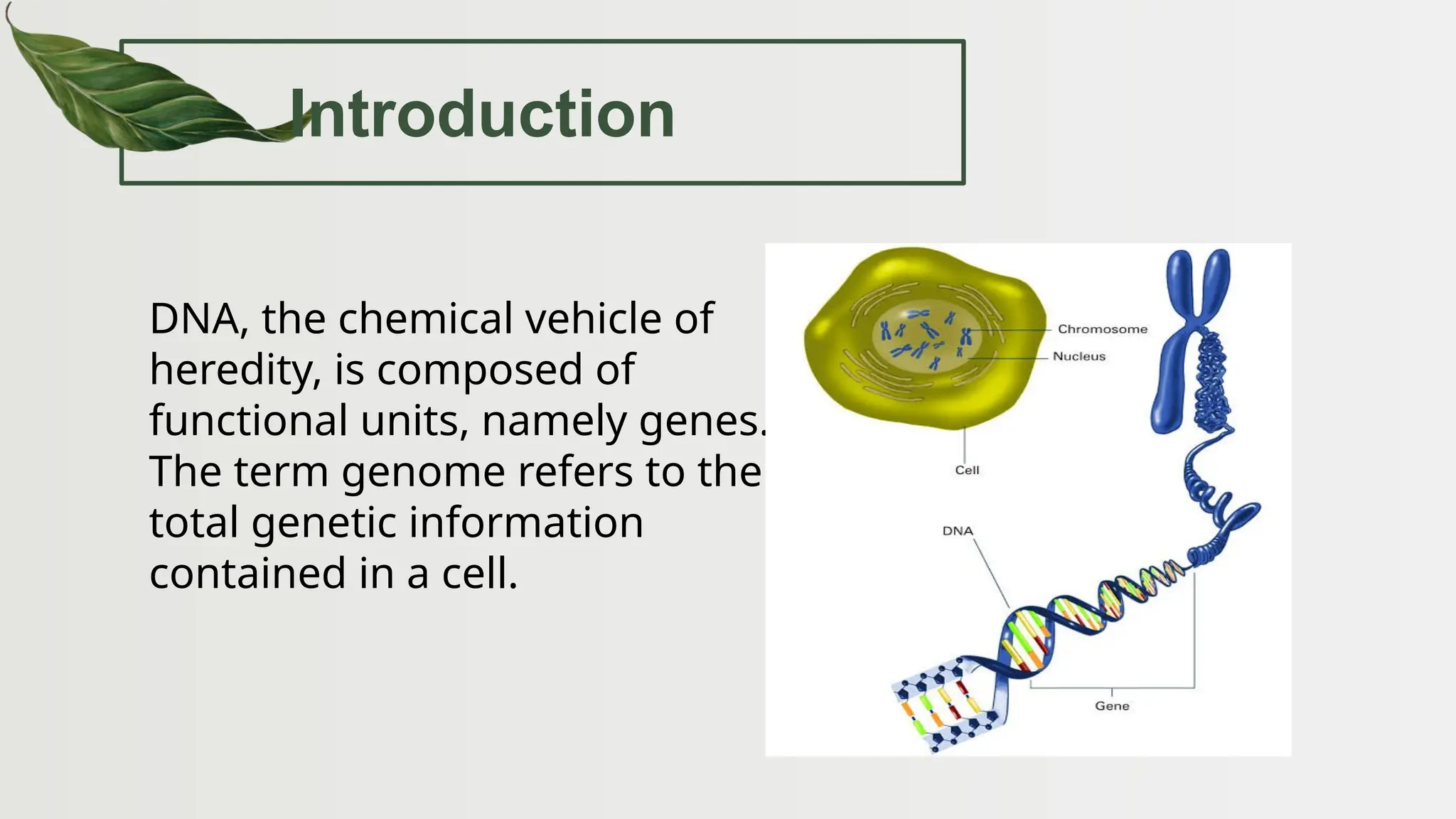
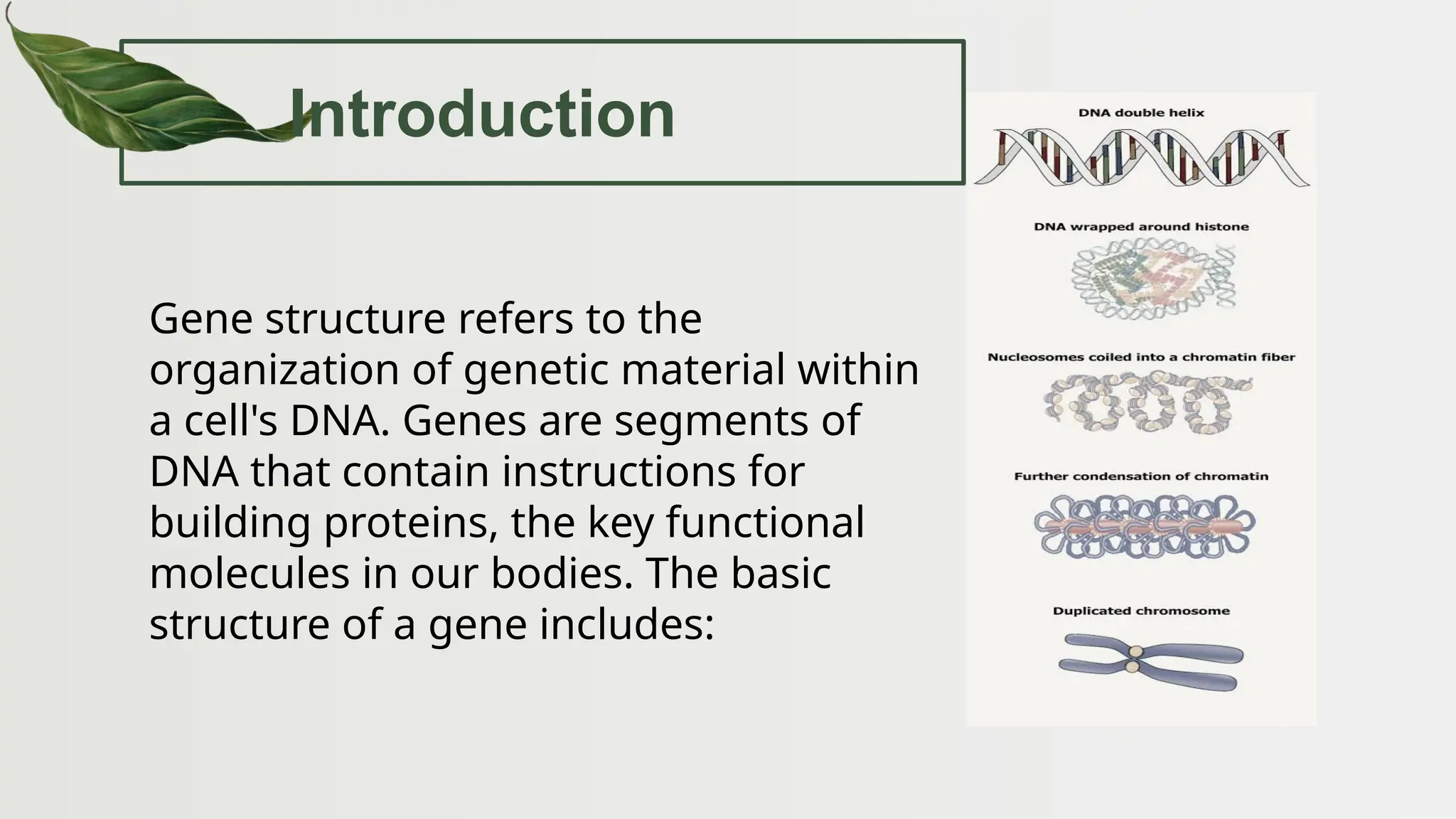
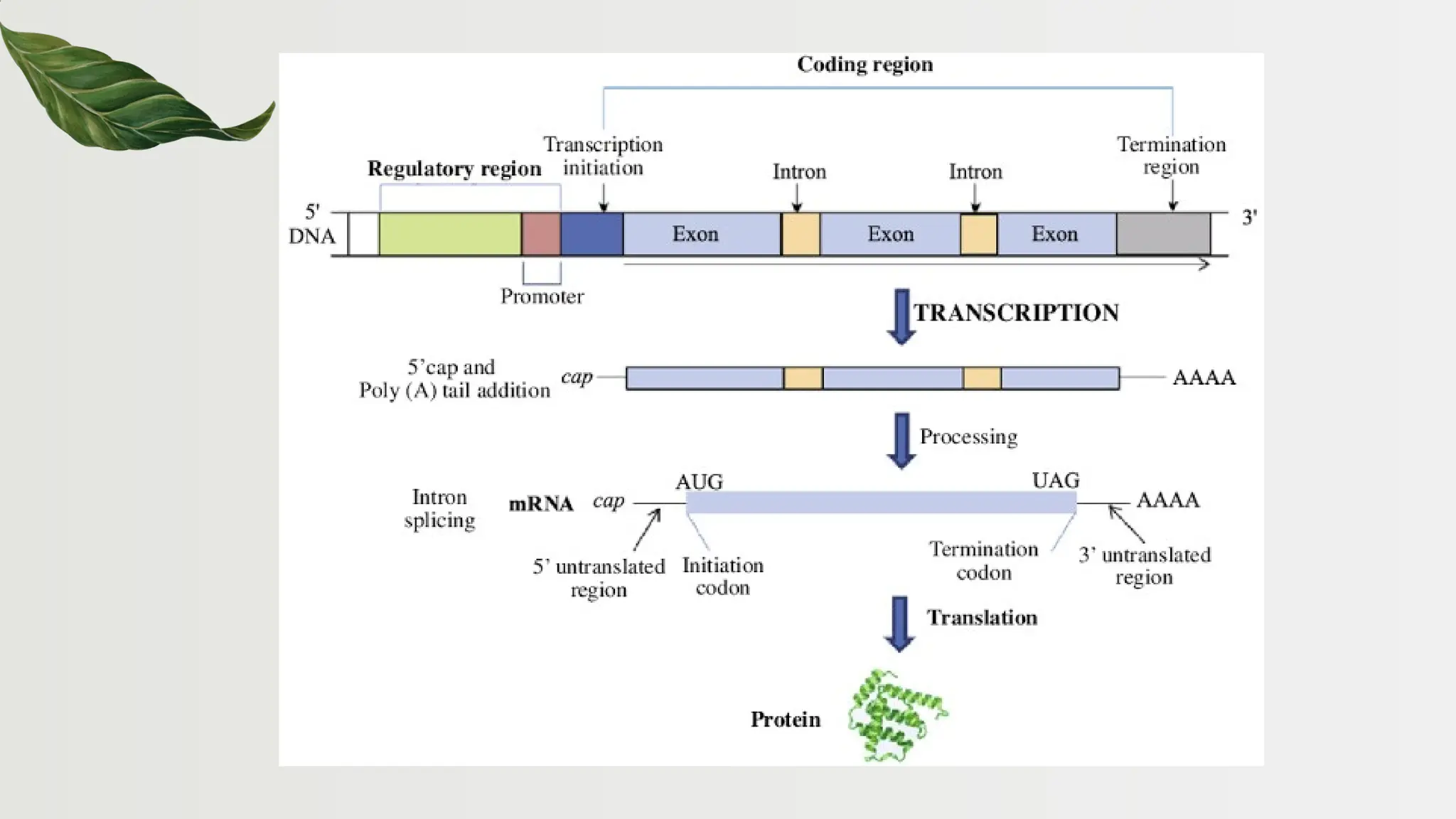
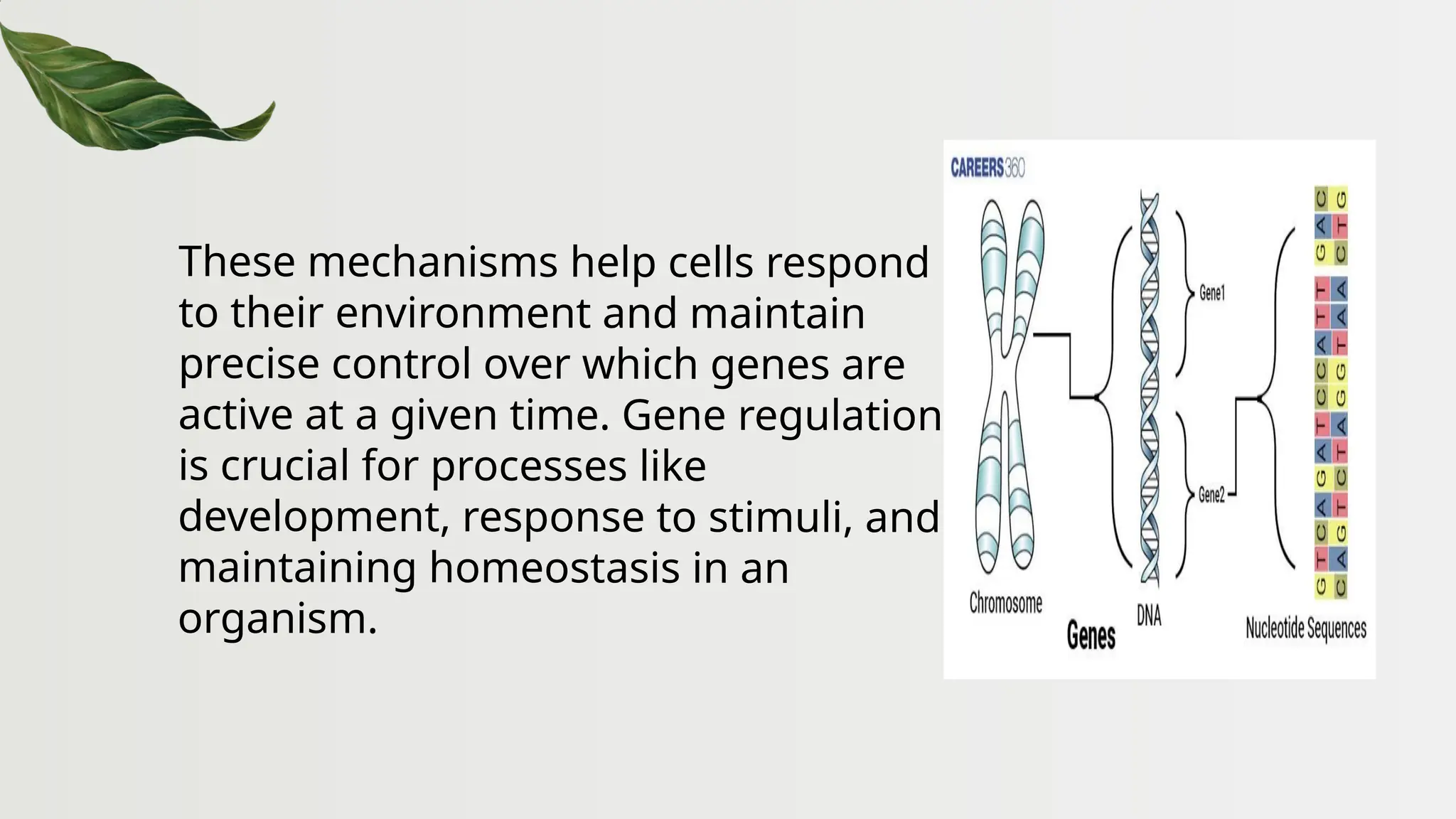
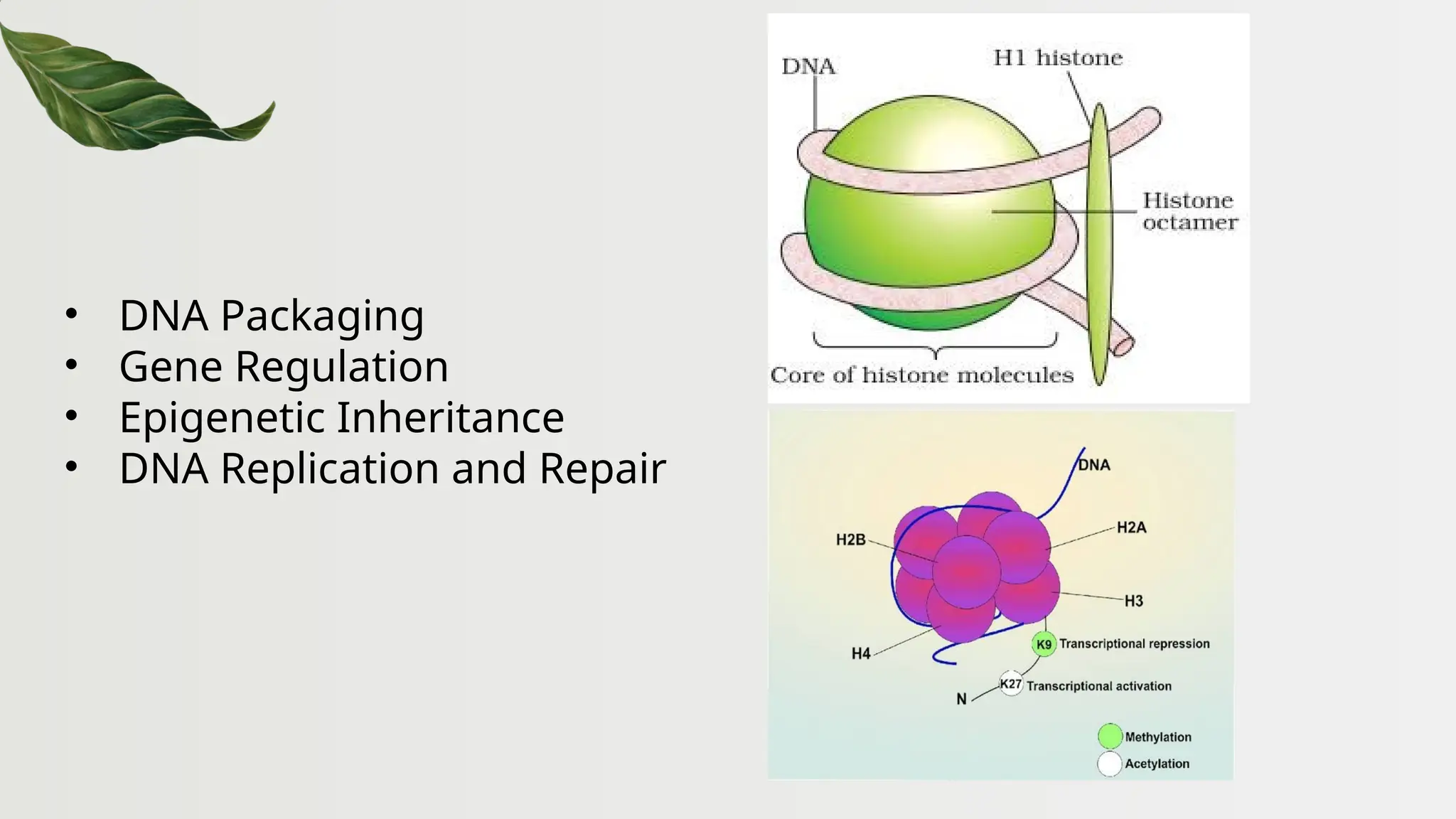
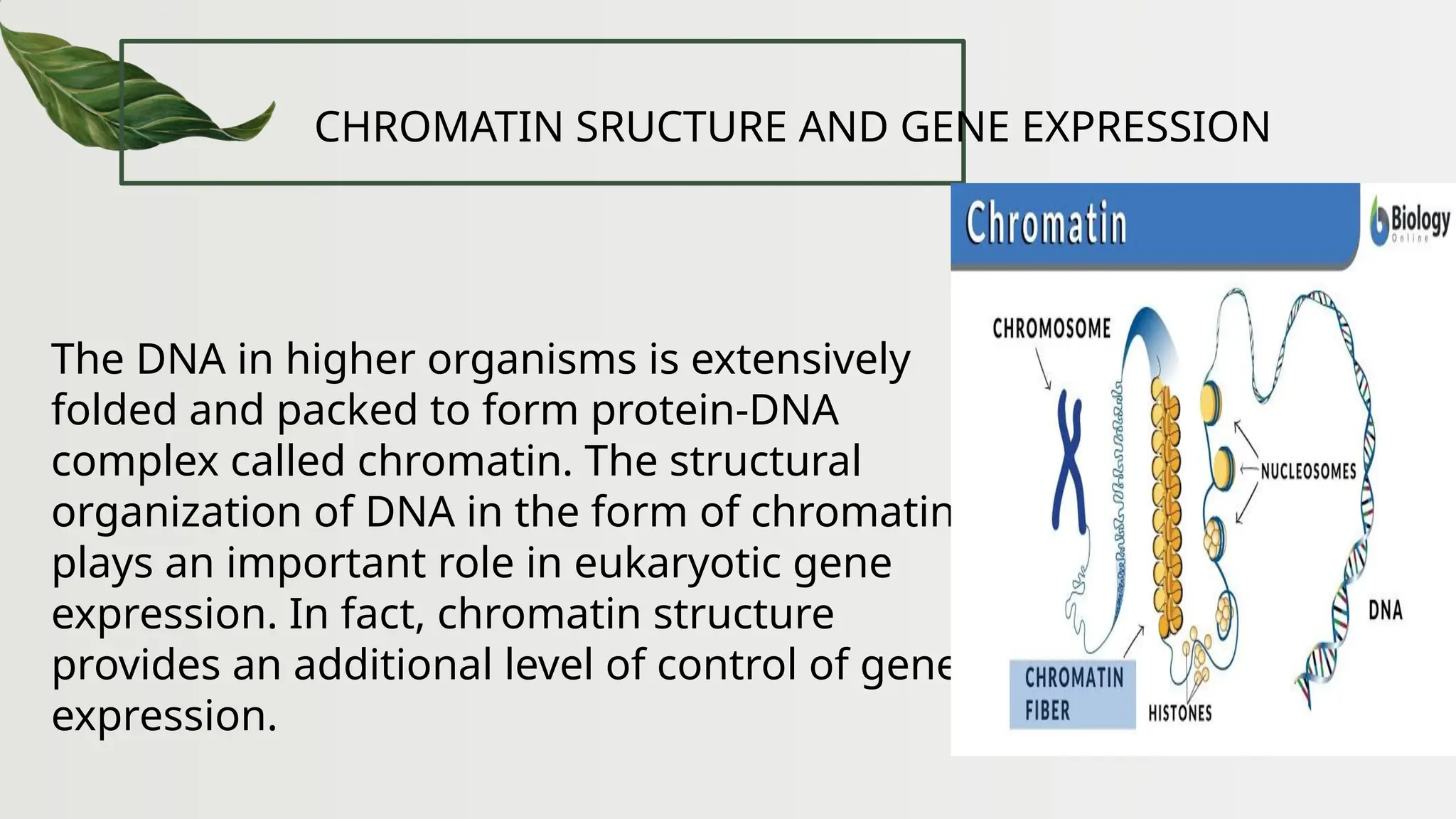
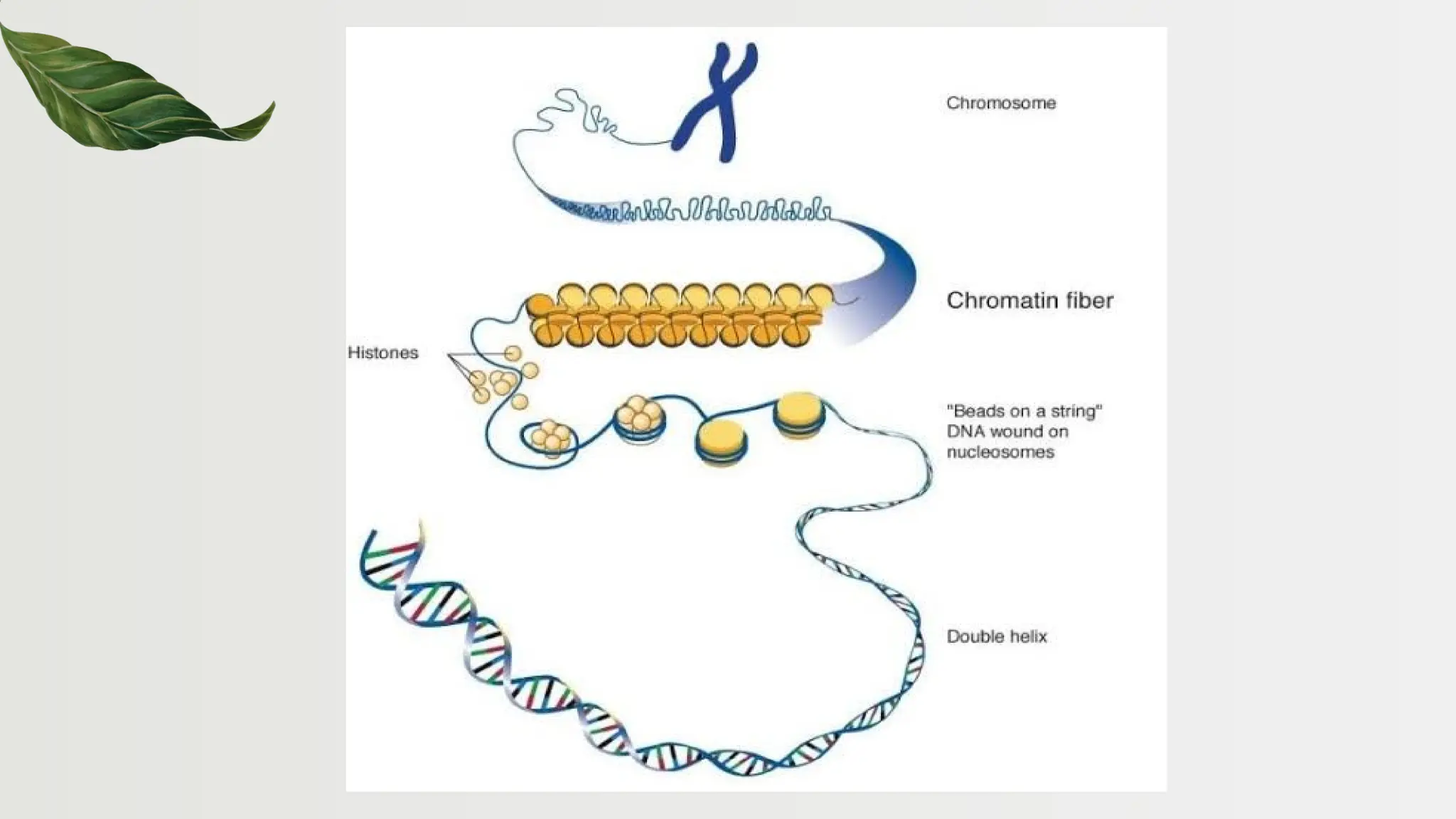
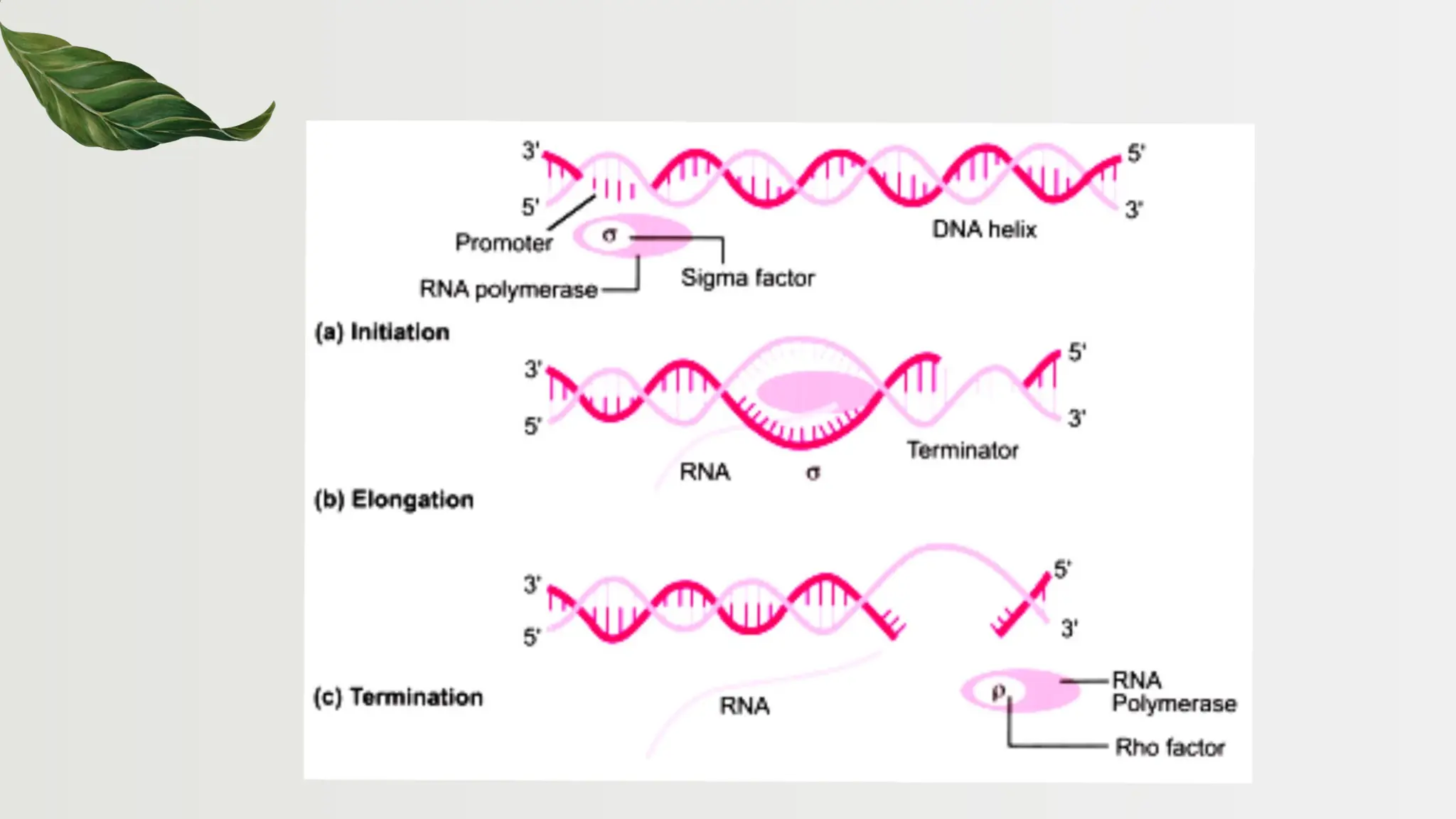
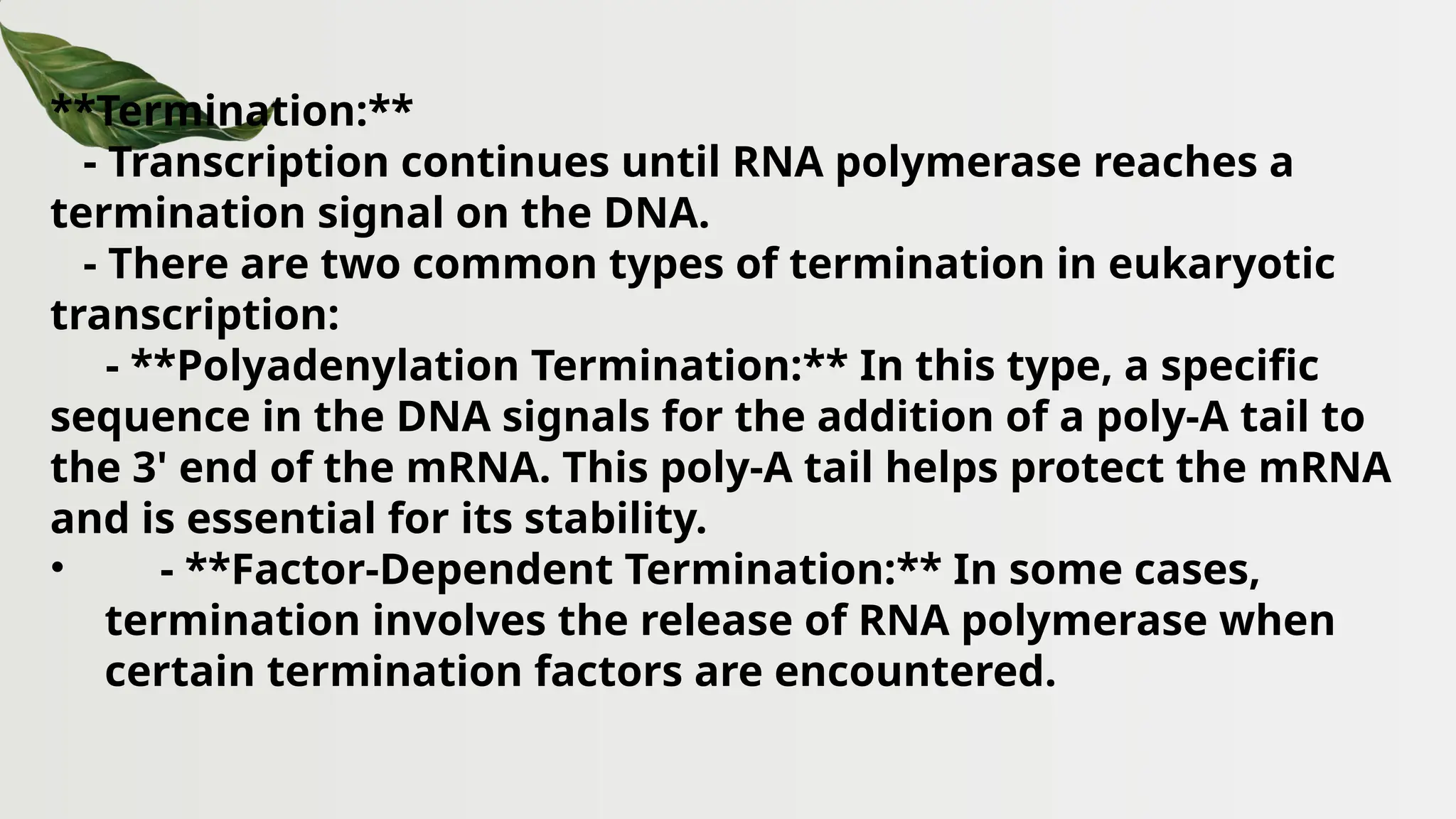
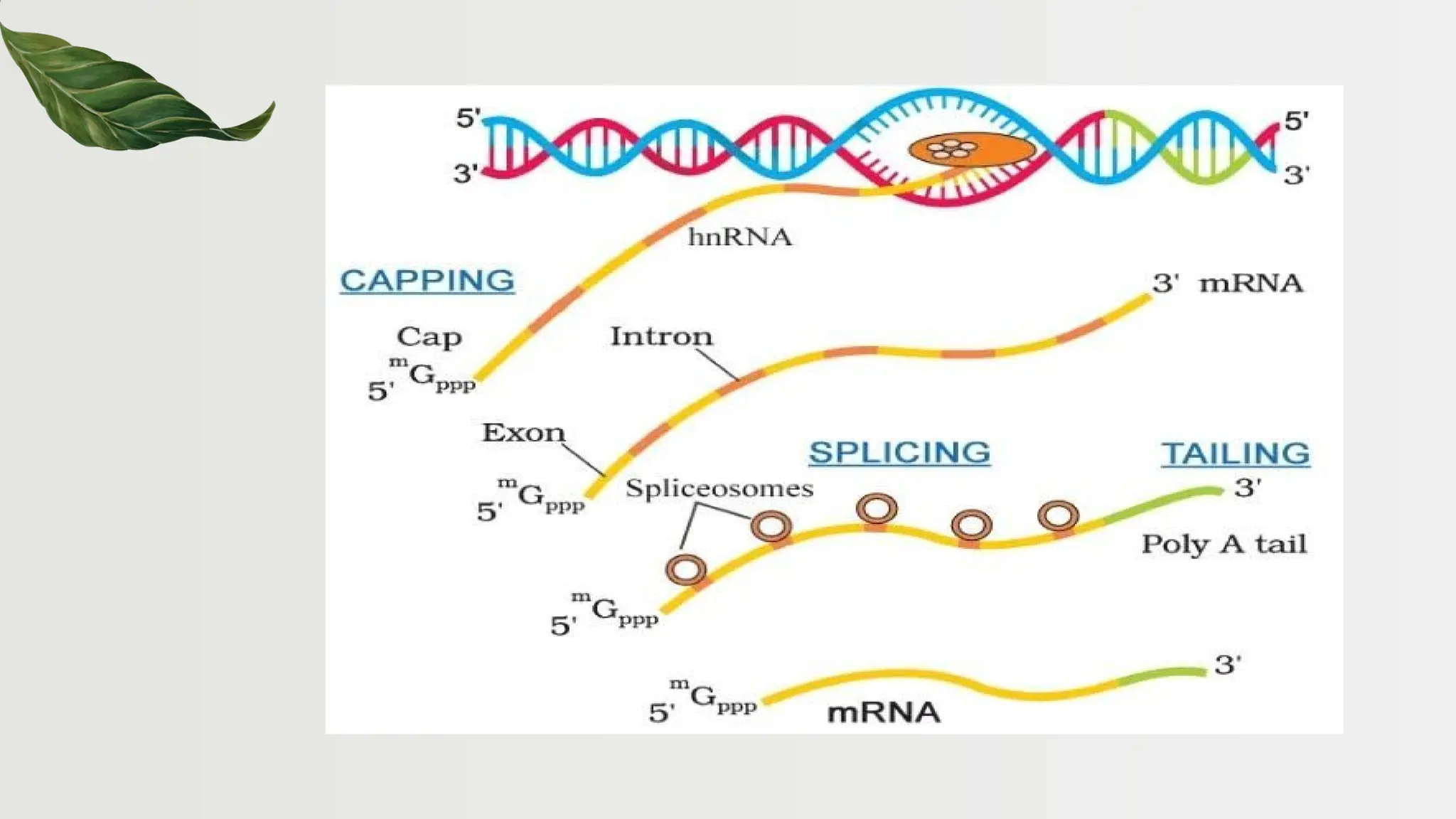
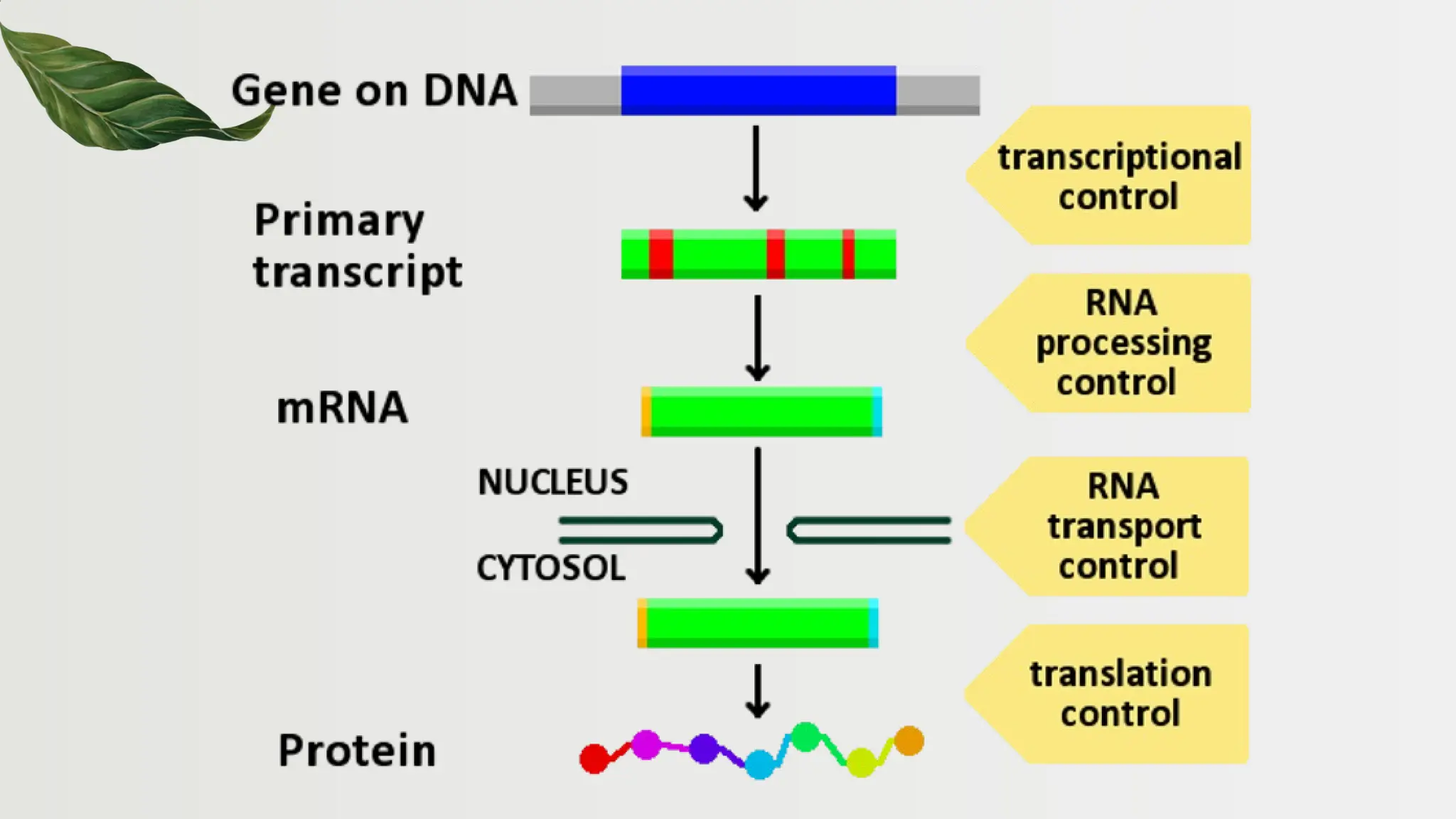
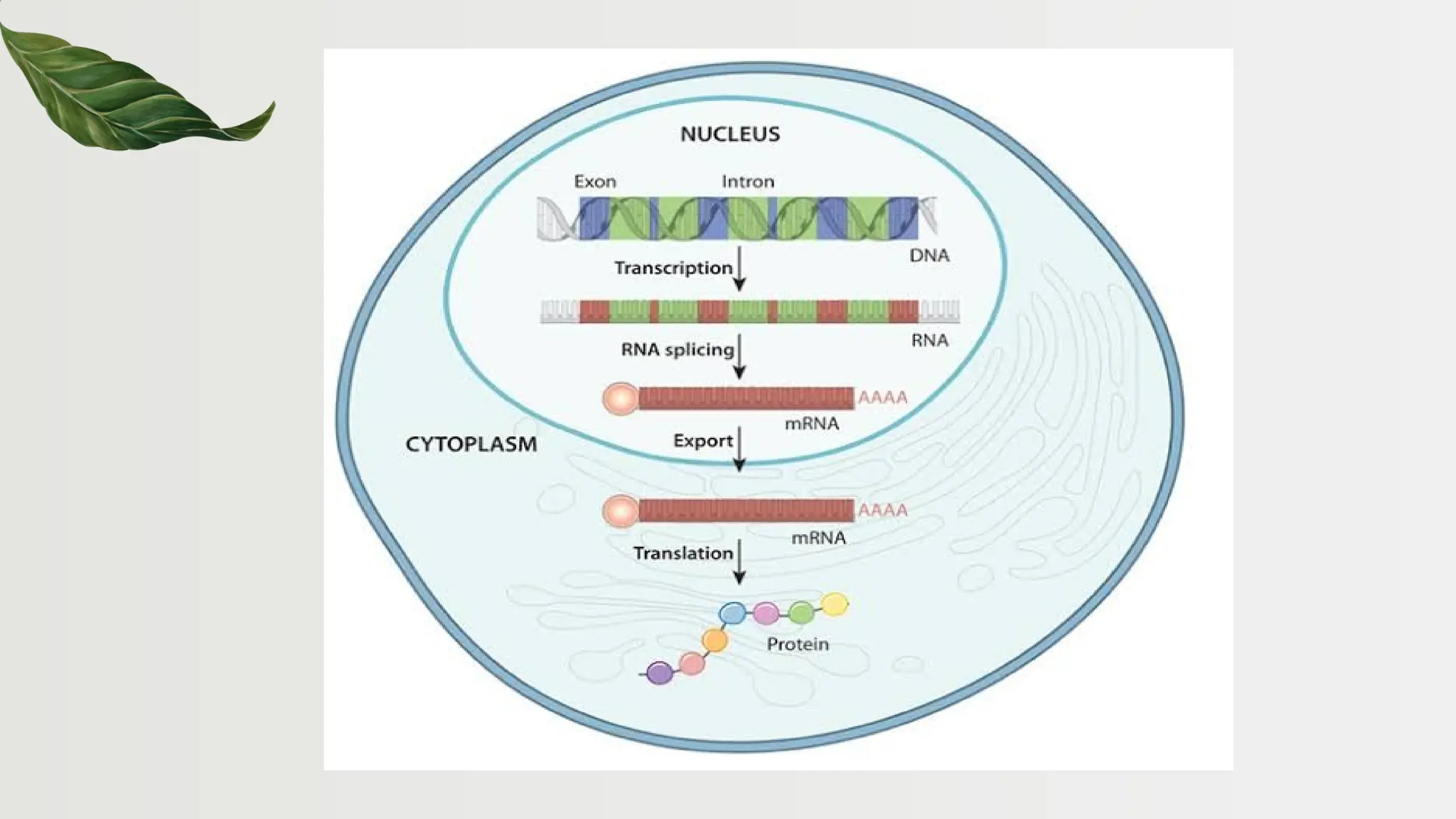
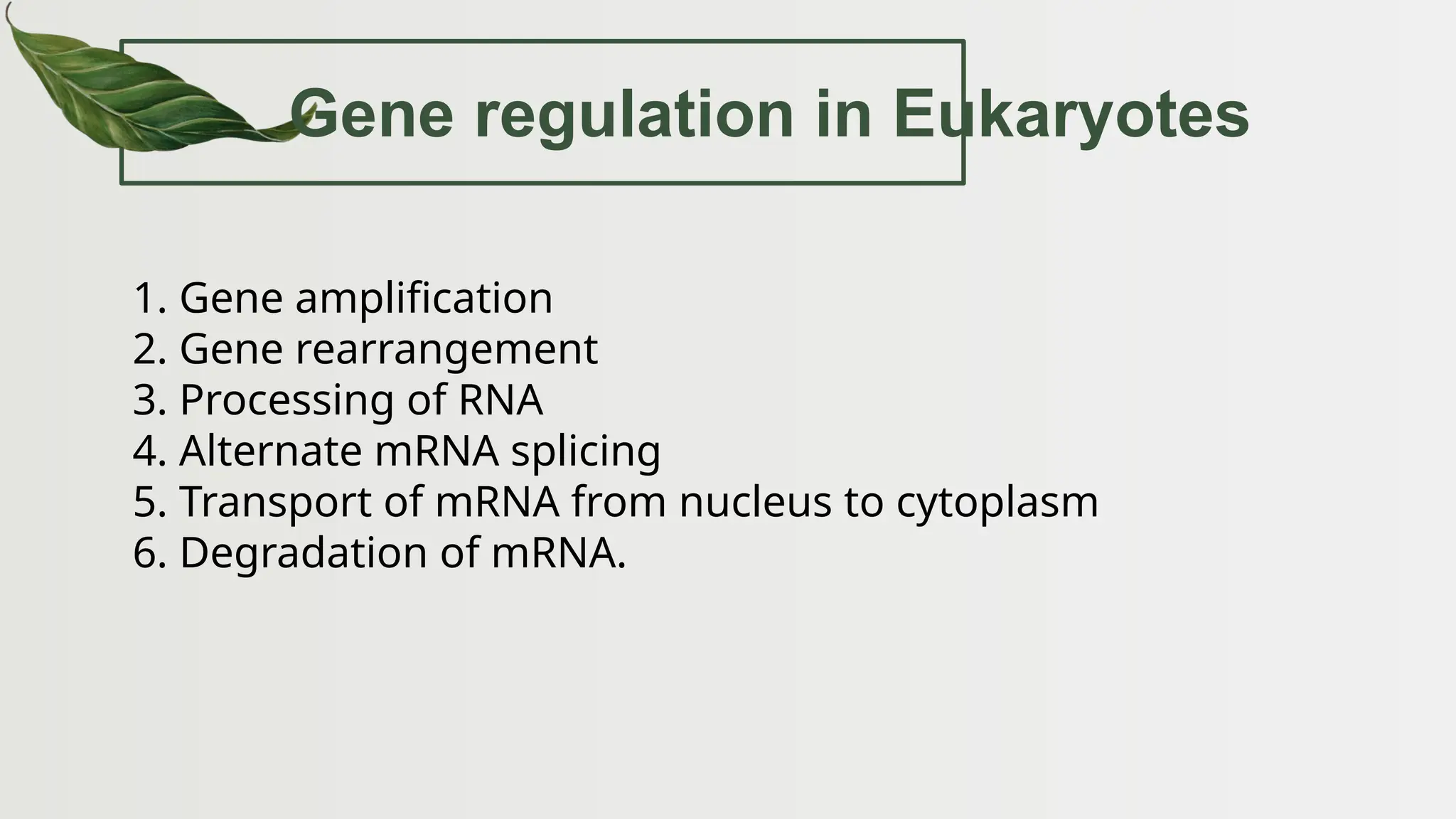
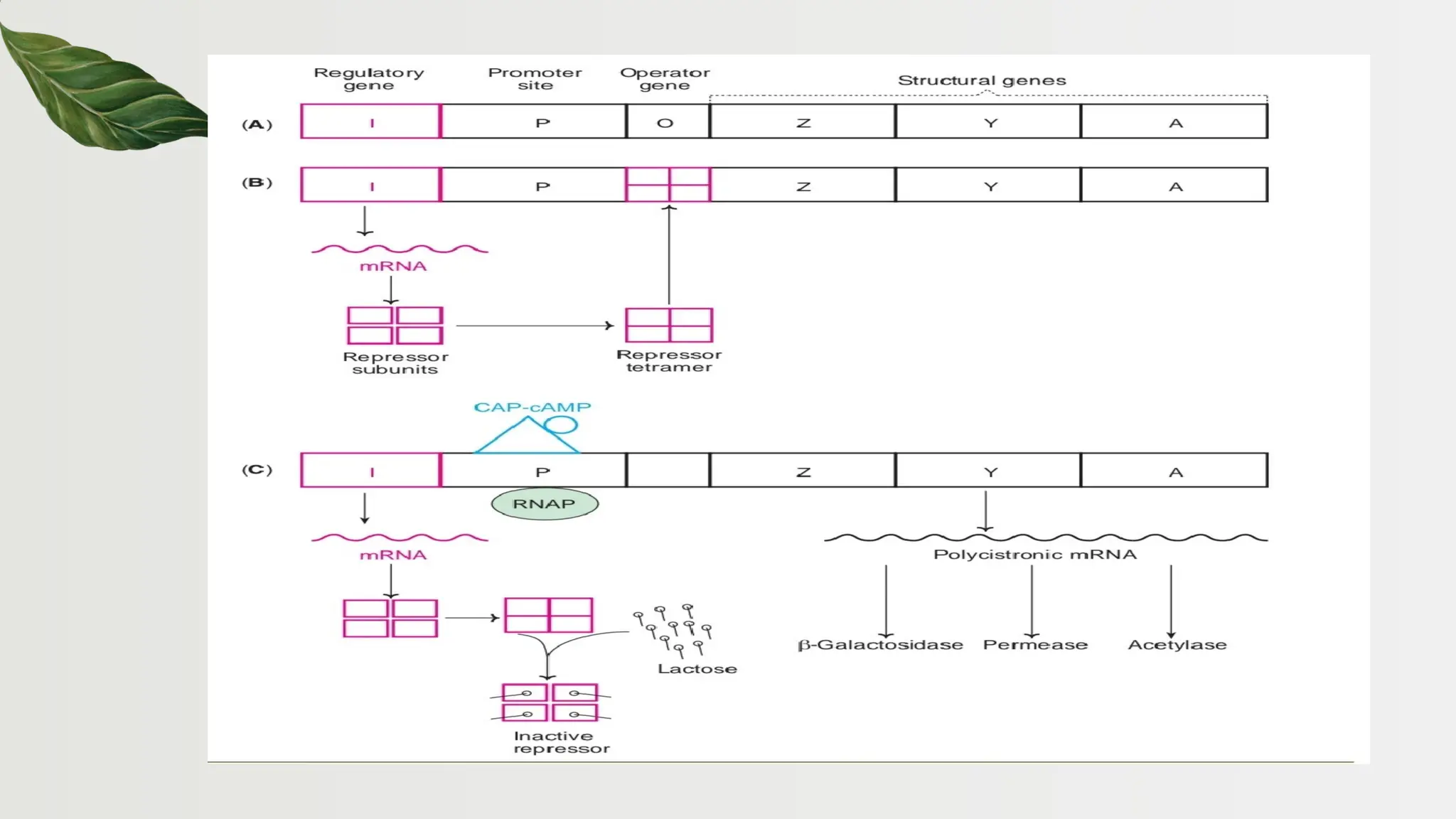
The document discusses the structure and regulation of genes, explaining the gene expression processes in both eukaryotes and prokaryotes. It covers key components such as DNA organization, transcription, RNA processing, and gene regulation mechanisms, emphasizing the importance of histones and regulatory proteins in gene expression control. Additionally, the document highlights the specific mechanisms of gene regulation in bacteria through operons, particularly the lac operon, and how these processes are essential for cellular function and response to environmental changes.