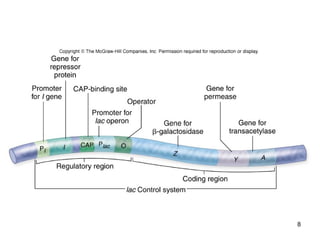
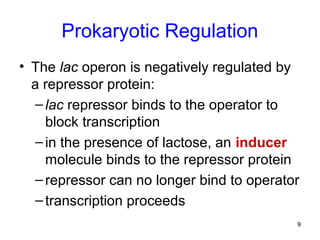
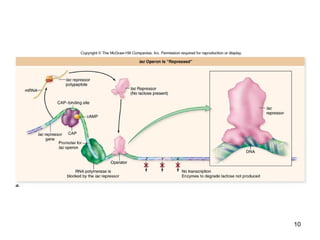
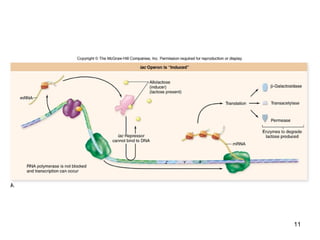
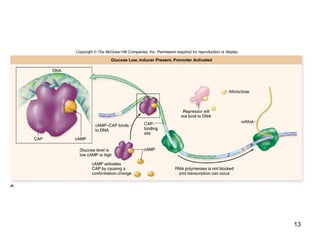
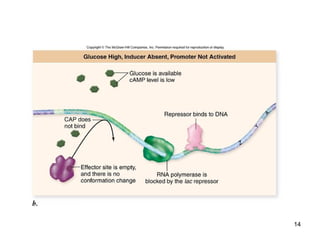
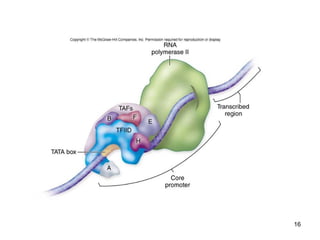
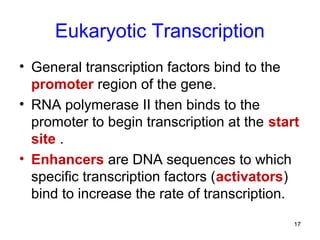
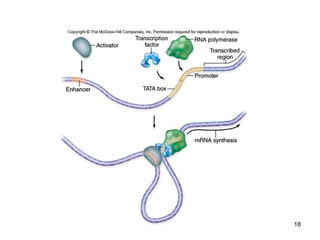
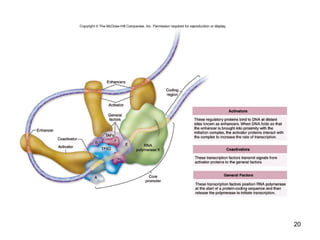
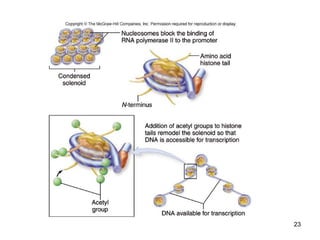
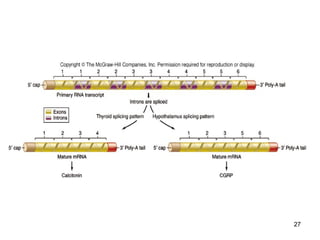
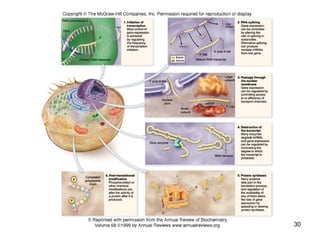
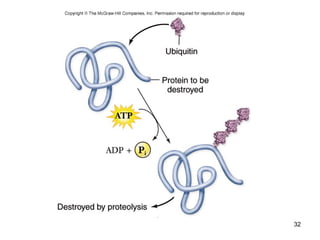
This document discusses the control of gene expression in prokaryotes and eukaryotes. In prokaryotes, genes are organized into operons and expressed through positive or negative transcriptional control when needed for metabolic pathways. The lac operon is negatively regulated by the lac repressor. In eukaryotes, general transcription factors and specific transcription factors regulate transcription, and chromatin structure and post-transcriptional mechanisms like alternative splicing further control gene expression.