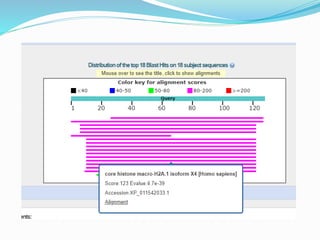
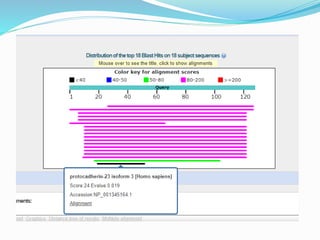
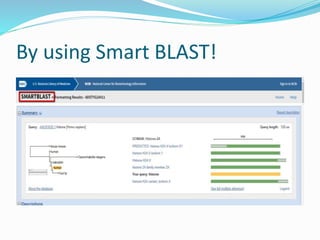
The Basic Local Alignment Search Tool (BLAST) is a pivotal resource in bioinformatics for comparing gene and protein sequences, allowing for faster local alignment searches than traditional dynamic programming methods. It supports various programs like BLASTn and BLASTp for nucleotide and protein comparisons, along with advanced functionalities for identifying relationships among sequences. BLAST is typically used for identifying species, locating protein domains, establishing phylogenies, and mapping DNA sequences.