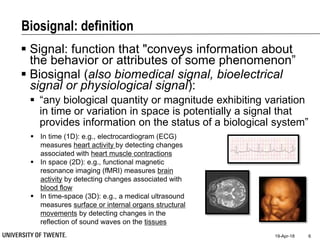
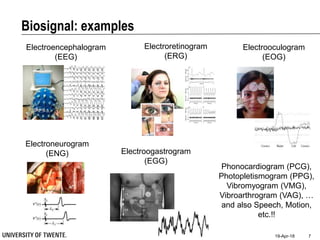
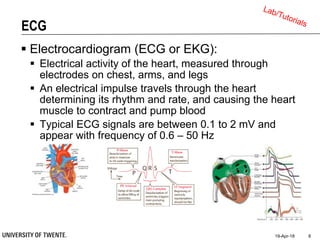
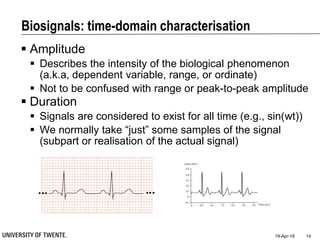
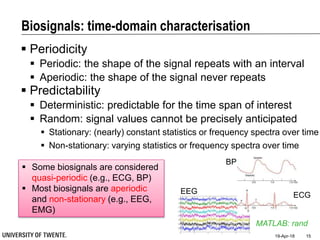
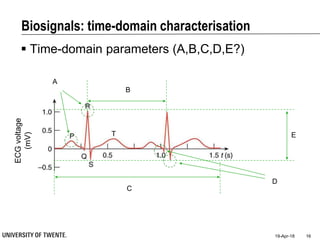
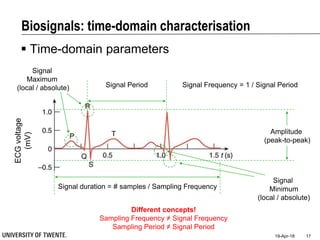
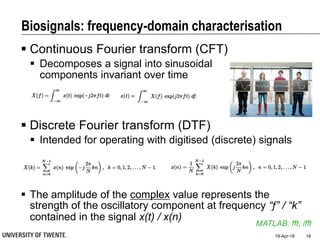
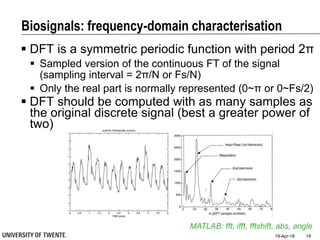
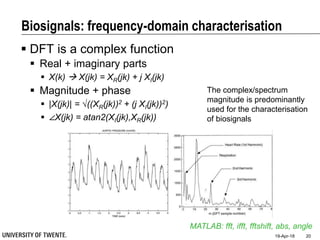
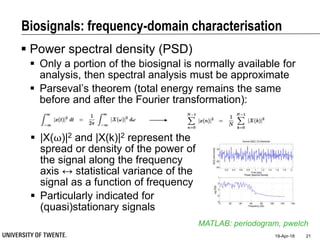
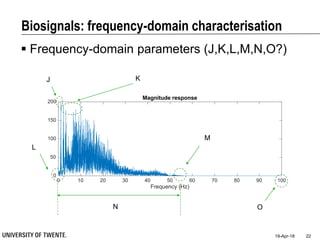
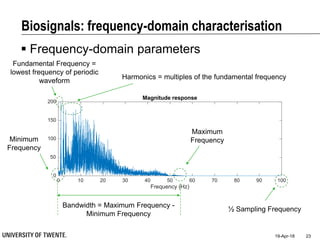
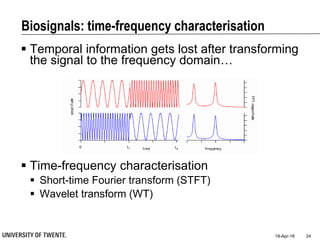
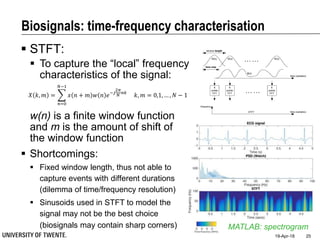
This document discusses biosignal processing and covers the following key points in 3 sentences:
It provides an overview of biosignal processing techniques including filtering to remove artifacts, event detection, and compression. It defines biosignals and gives examples like ECG and EMG. The document outlines topics like characterizing biosignals in the time and frequency domains, and techniques for time-frequency analysis like short-time Fourier transform and wavelet transform.

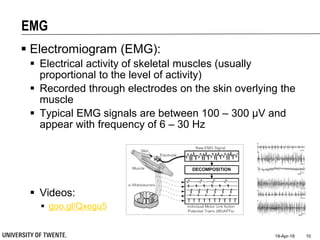
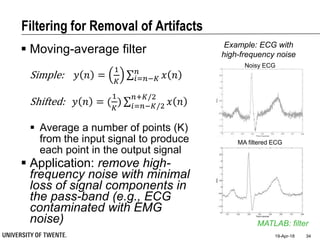
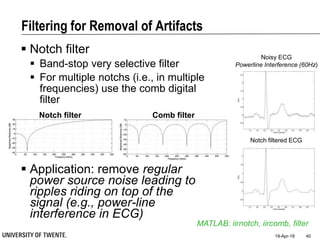
![Filtering for Removal of Artifacts
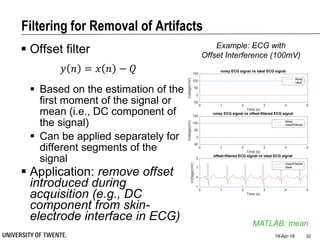
Median filter
Ranks from lowest value to
highest value and picks the
middle one:
e.g., median of [3, 3, 5, 9, 11] is 5
Good for rejecting certain
types of noise in which some
samples take extreme values
(outliers)
Application: regular power
source leading to ripples
riding on top of the signal
(e.g., power source
interference in ECG)
19-Apr-18 35
MATLAB: medfilt1
0 1 2 3 4 5
Time (s)
-1
0
1
2
ECG noisy signal and ideal signal
Noisy
Ideal
0 1 2 3 4 5
Time (s)
-1
0
1
2
ECG noisy signal and median-filtered signal
Noisy
medFiltered
0 1 2 3 4 5
Time (s)
-1
0
1
2
Median-filtered signal and ideal signal
medFiltered
Ideal
Example: ECG with
Powerline Interference (50Hz)](https://image.slidesharecdn.com/biosignalprocessing-180419100353/85/Biosignal-Processing-35-320.jpg)
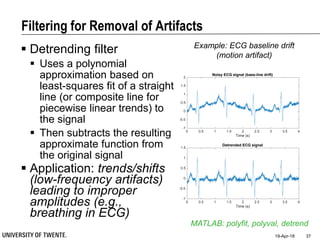
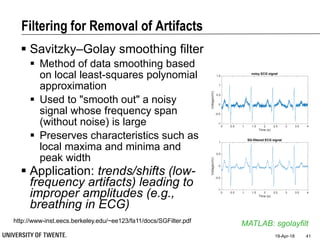
![Filtering for Removal of Artifacts
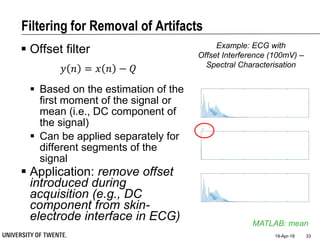
Median filter
Ranks from lowest value to
highest value and picks the
middle one:
e.g., median of [3, 3, 5, 9, 11] is 5
Good for rejecting certain
types of noise in which some
samples take extreme values
(outliers)
Application: regular power
source leading to ripples
riding on top of the signal
(e.g., power source
interference in ECG)
19-Apr-18 36
MATLAB: medfilt1
0 10 20 30 40 50 60 70 80 90 100
Frequency (Hz)
0
500
1000
PSD (ECG ideal)
0 10 20 30 40 50 60 70 80 90 100
Frequency (Hz)
0
500
1000
1500
PSD (ECG noisy)
0 10 20 30 40 50 60 70 80 90 100
Frequency (Hz)
0
500
1000
PSD (ECG filtered)
Example: ECG with
Powerline Interference (50Hz) –
Spectral Characterisation](https://image.slidesharecdn.com/biosignalprocessing-180419100353/85/Biosignal-Processing-36-320.jpg)


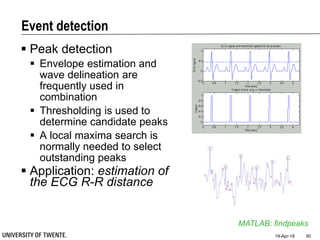
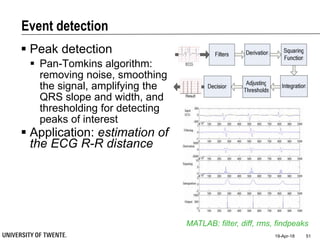
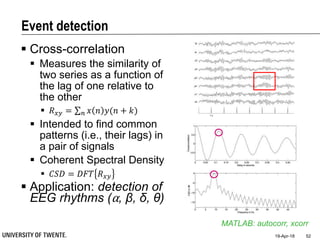
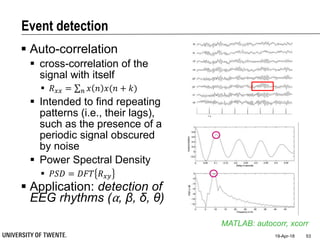

![References
Digital Signal Processing
Principal reference:
Proakis, J. G., & Manolakis, D. G. Digital Signal Processing. Principles,
Algorithms, and Applications. New Jersey: Prentice-Hall, 1996
Other references:
Hayes, M. Statistical Digital Signal Proccessing and Modeling. New York: John
Wiley & Sons, 1996
[OPEN BOOK] Smith, Steven W., The Scientist and Engineer's Guide to Digital
Signal Processing, http://www.dspguide.com/pdfbook.htm
[OPEN BOOK] Prandoni, P., Martin, V., Signal Processing for Communications,
http://www.sp4comm.org/docs/sp4comm_corrected.pdf
19-Apr-18 58](https://image.slidesharecdn.com/biosignalprocessing-180419100353/85/Biosignal-Processing-58-320.jpg)
