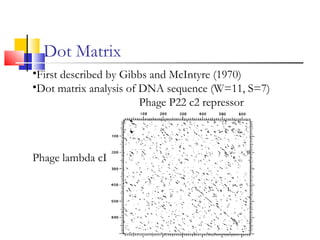
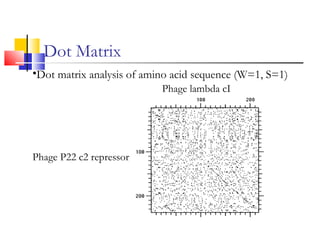
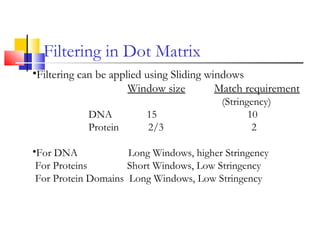
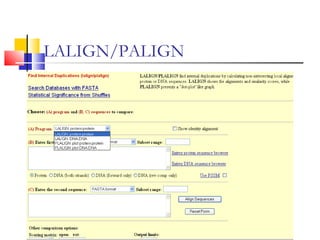
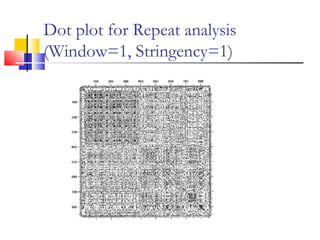
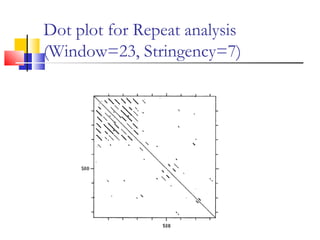
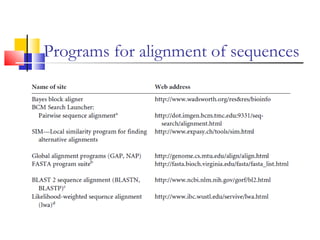
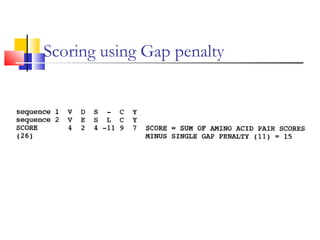
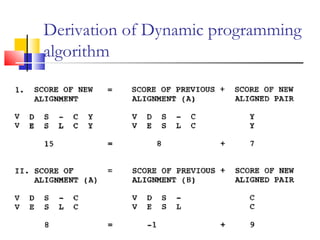
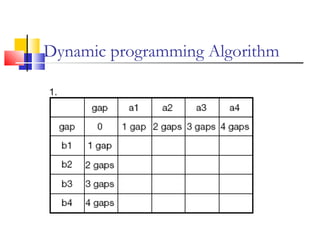
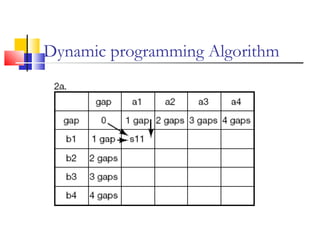
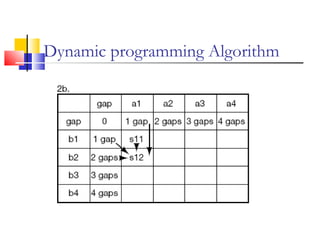
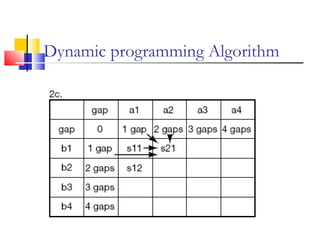
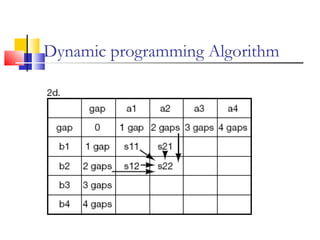
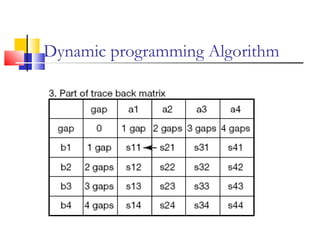
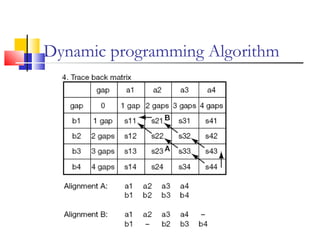
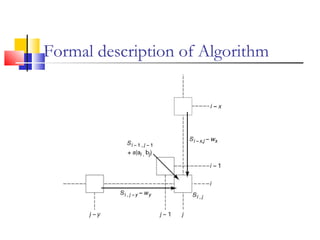
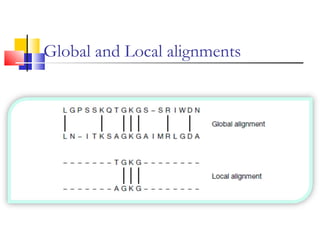
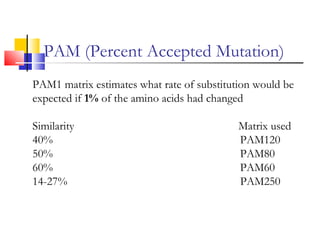
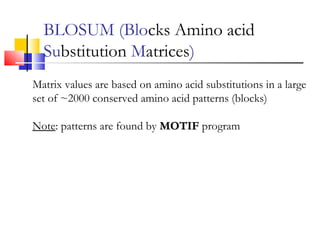
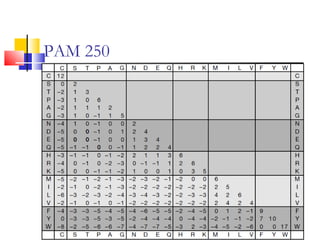
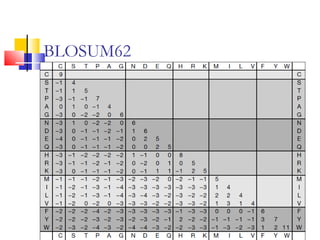
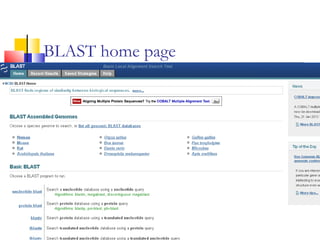
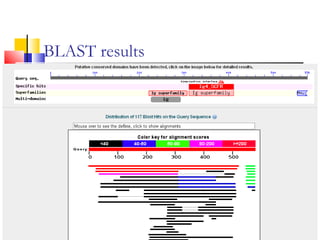
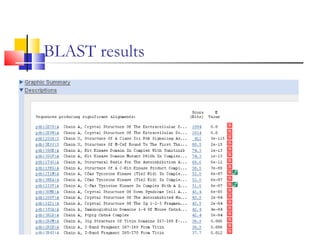
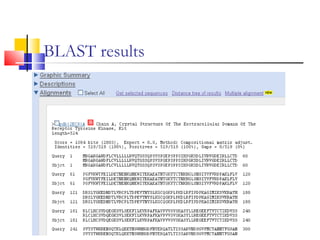
This document discusses various statistical methods used in bioinformatics, including dot matrix analysis and dynamic programming algorithms. Dot matrix analysis can be used to compare DNA or protein sequences visually and look for repeats or alignments. Dynamic programming algorithms like those used in BLAST are commonly used to generate optimal global and local sequence alignments by calculating alignment scores based on substitution matrices like PAM and BLOSUM.