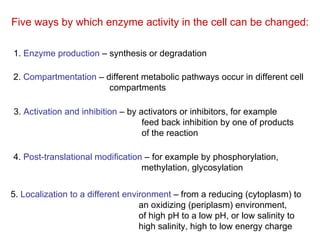
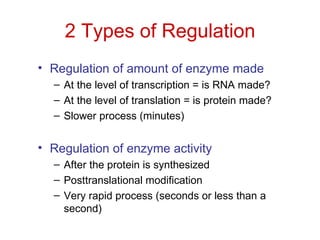
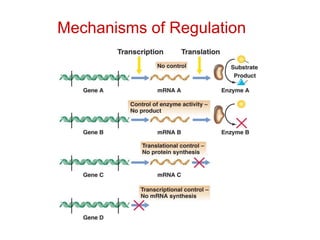
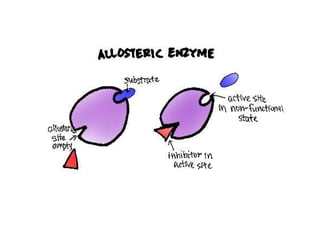
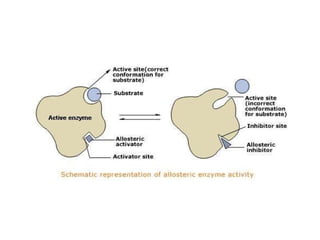
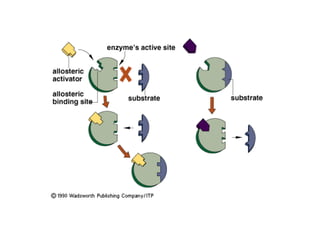
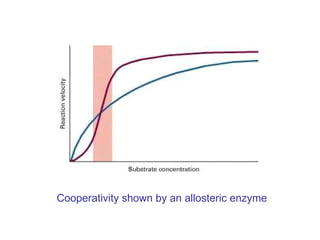
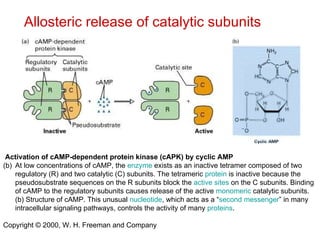
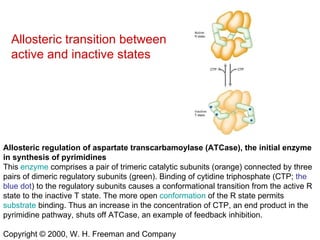
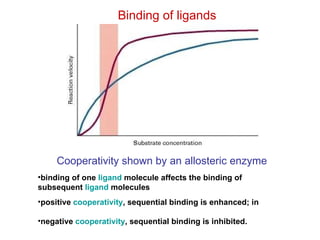
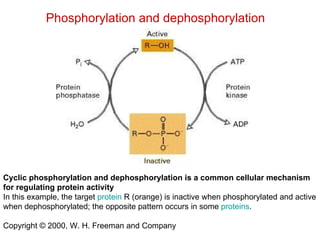
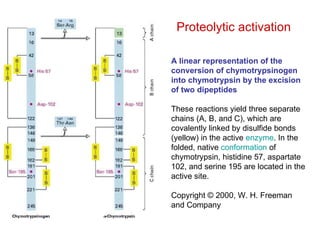
This document summarizes different ways that enzyme activity can be regulated in cells, including at the level of production, compartmentation, activation/inhibition, post-translational modification, and localization. It describes constitutive enzymes that are always active and regulated enzymes that are only active under certain conditions. Enzyme activity can be regulated at the level of transcription/translation or after protein synthesis via posttranslational modification. Mechanisms of regulation include feedback inhibition, allosteric regulation of activity based on substrate or product binding, phosphorylation/dephosphorylation, and proteolytic activation.