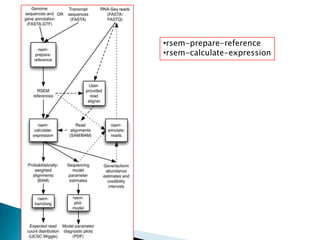
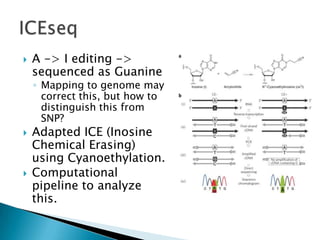
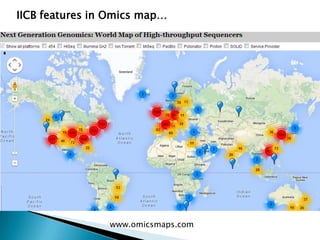
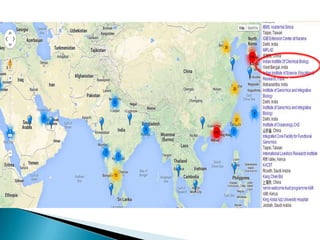
This document summarizes information from several genomics and bioinformatics research groups and projects. It discusses:
- The ENCODE project and its focus areas including databases, data mining, visualization, transcriptomics, alternative splicing, sequencing pipelines, comparative genomics, epigenomics, and population genomics.
- Tools and databases for variant analysis from the 1000 Genomes Project and FORGE Consortium.
- The Genome Modeling System from The Genome Institute at Washington University for analyzing TCGA, ICGC, 1000 Genomes, and PCGP data.
- Using RNA-seq technology to reveal the transcriptome and methods for isolating translated mRNA.
- Resources for analyzing Human Microbiome Project data





![
RNA-seq (RNA Sequencing), also called
"Whole Transcriptome Shotgun
Sequencing" [1] ("WTSS"), is a technology that
uses the capabilities of next-generation
sequencing to reveal a snapshot of RNA
presence and quantity from a genome at a
given moment in time.[2]](https://image.slidesharecdn.com/26nov2013seminar-131126065546-phpapp01/85/26-nov2013seminar-7-320.jpg)