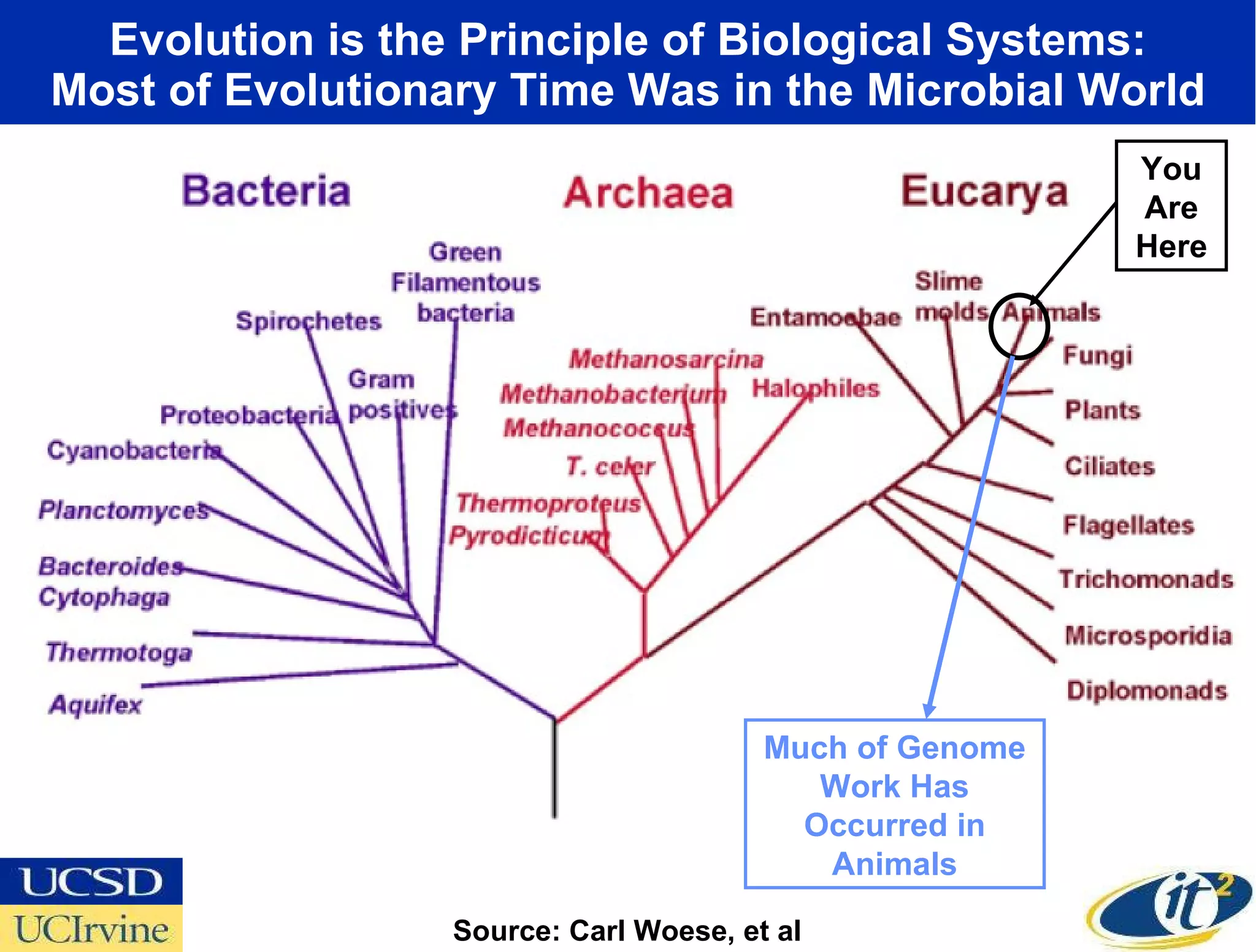
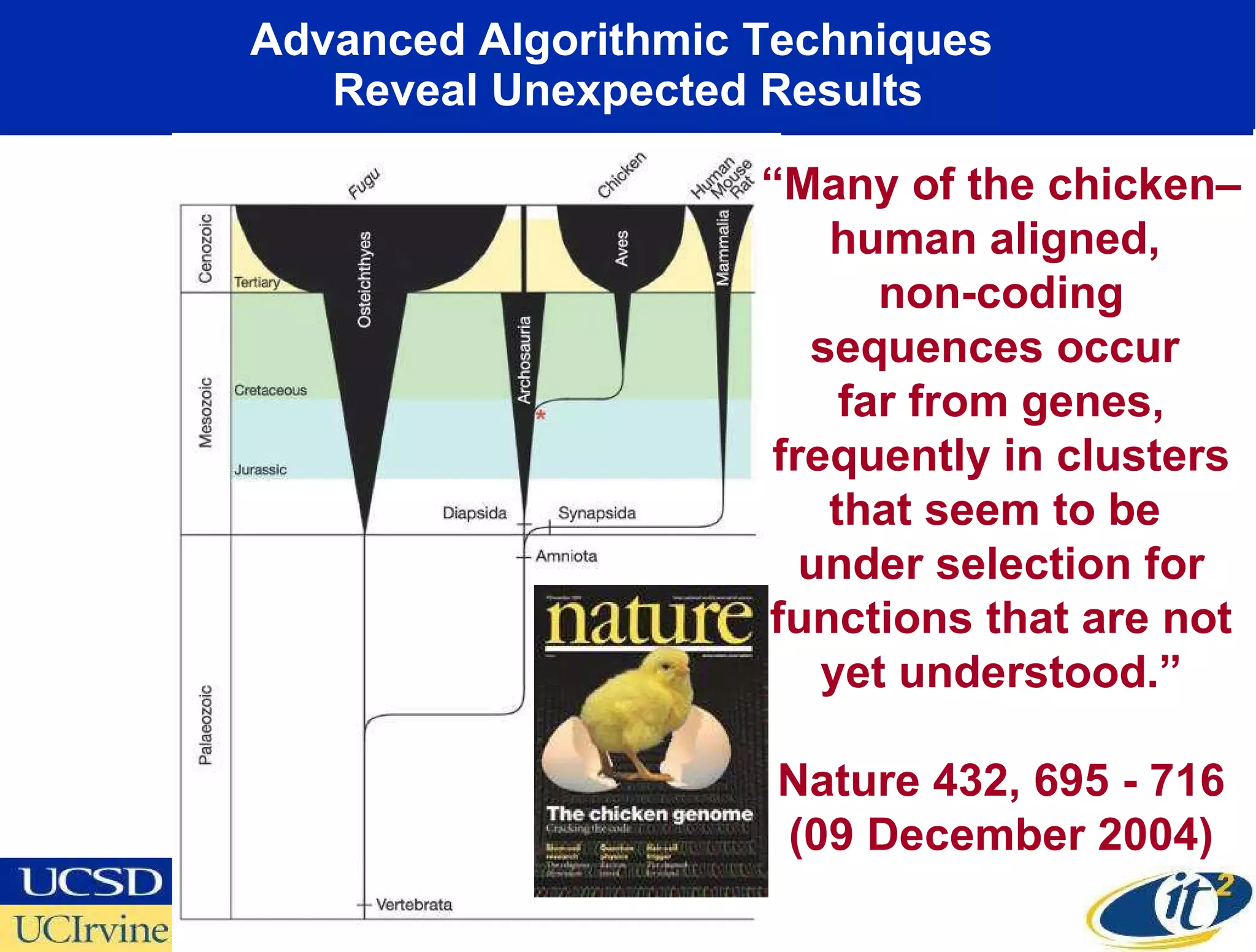
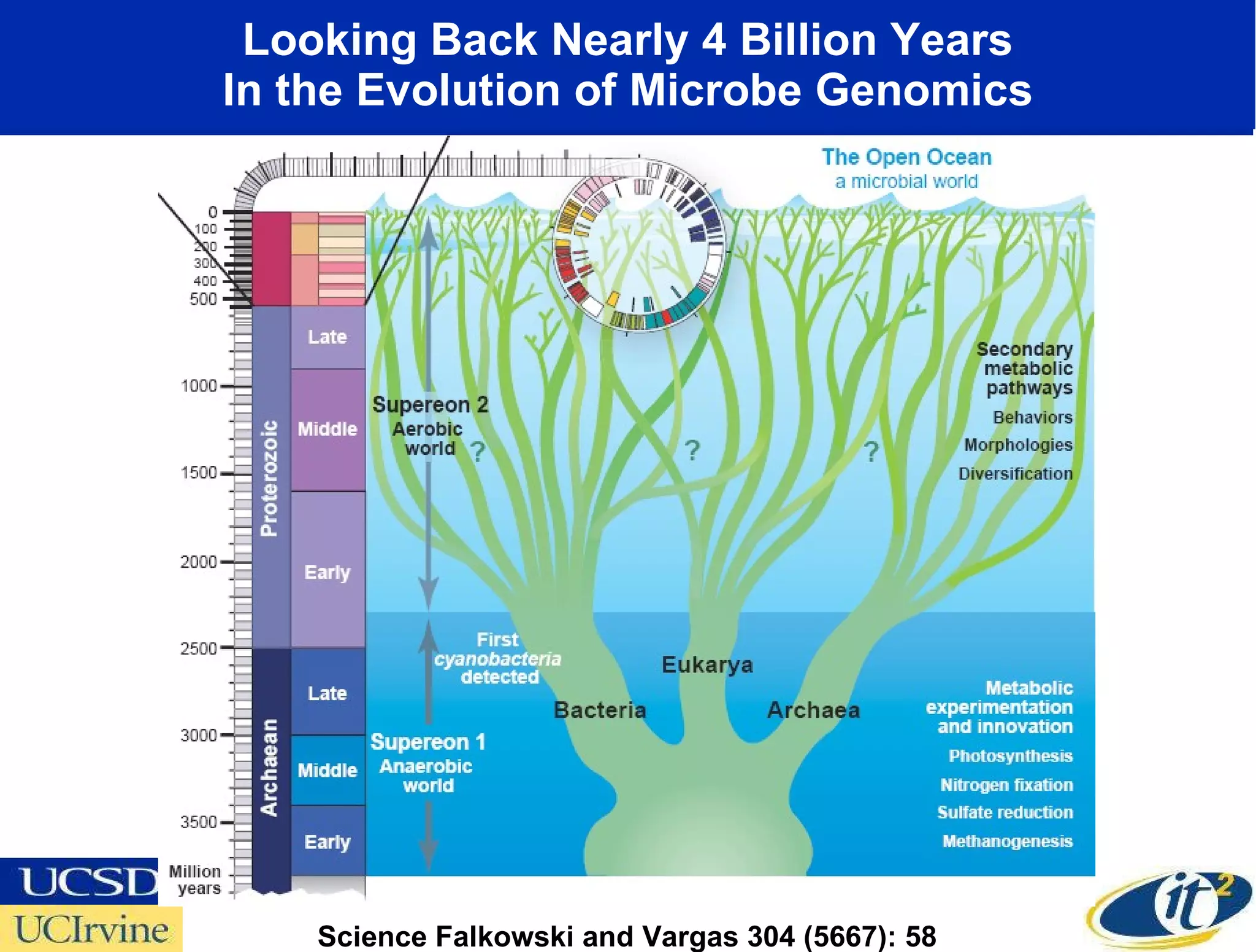
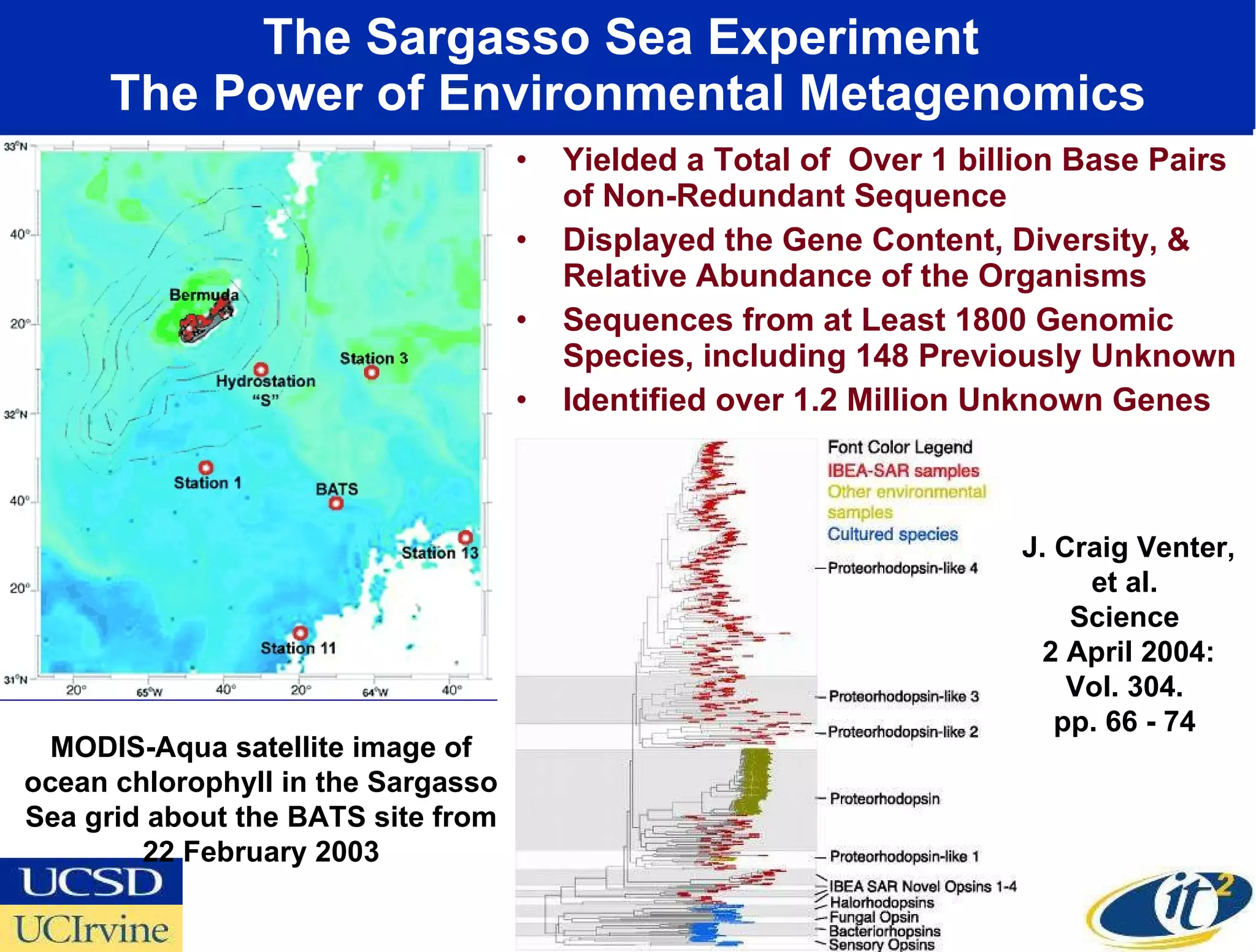
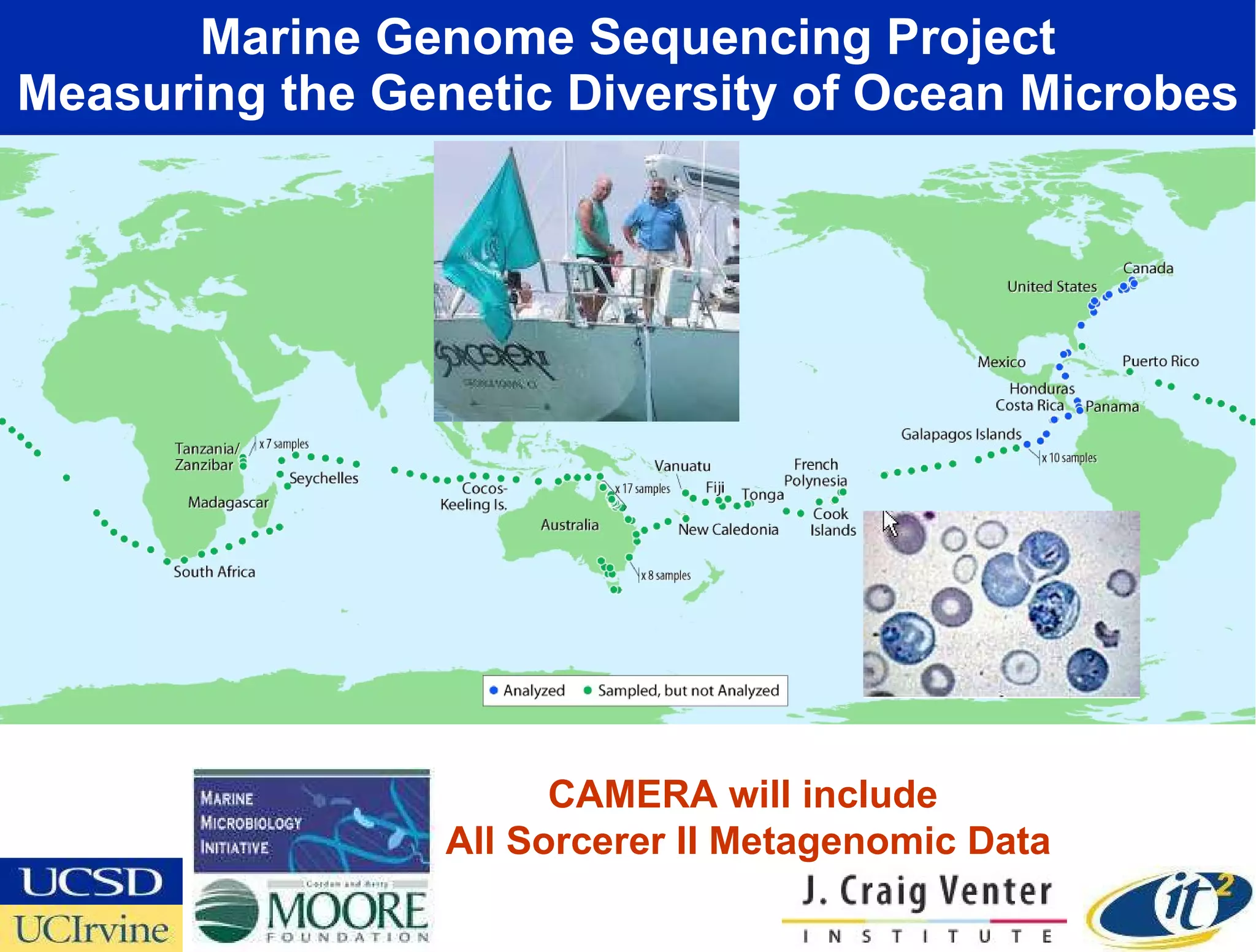
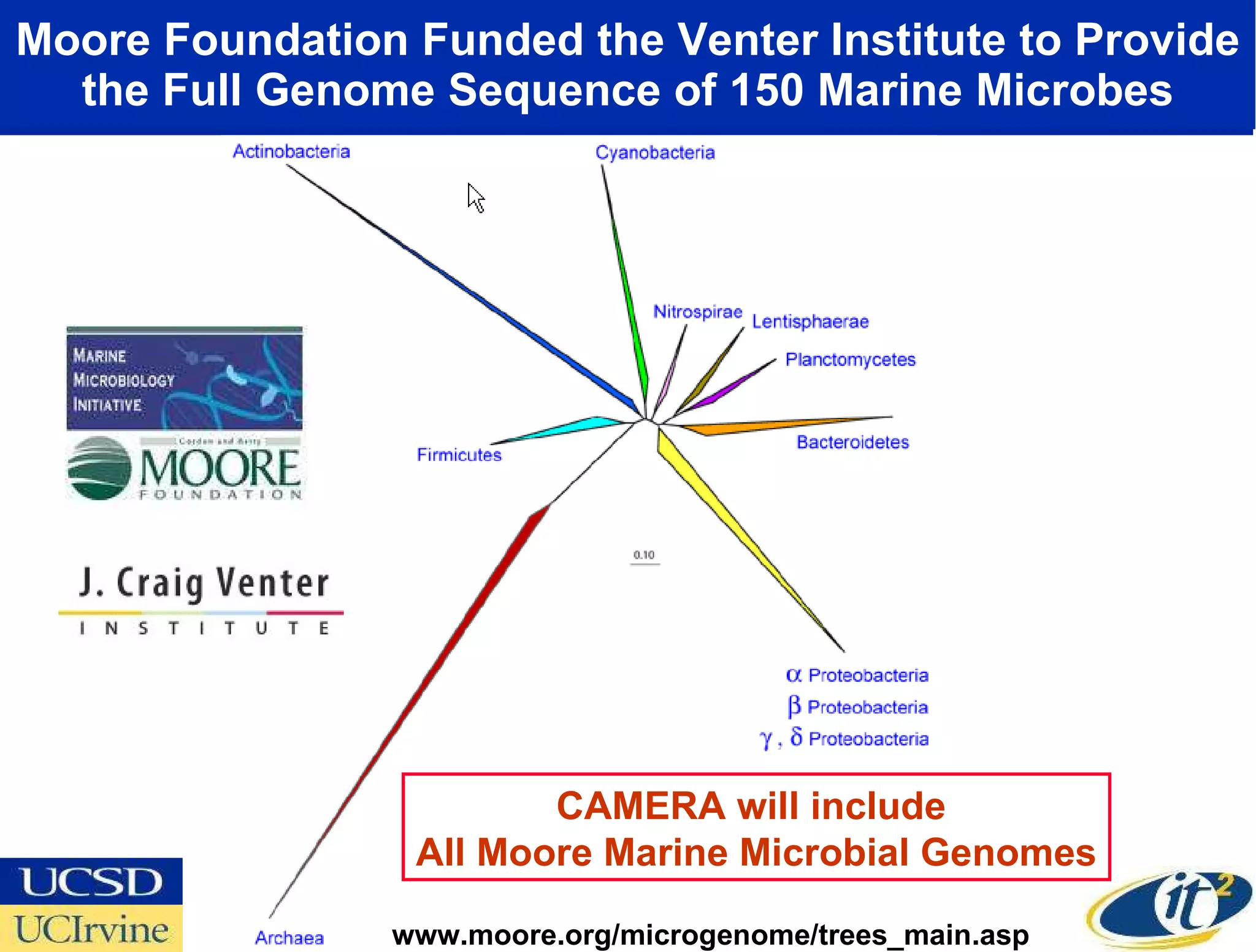
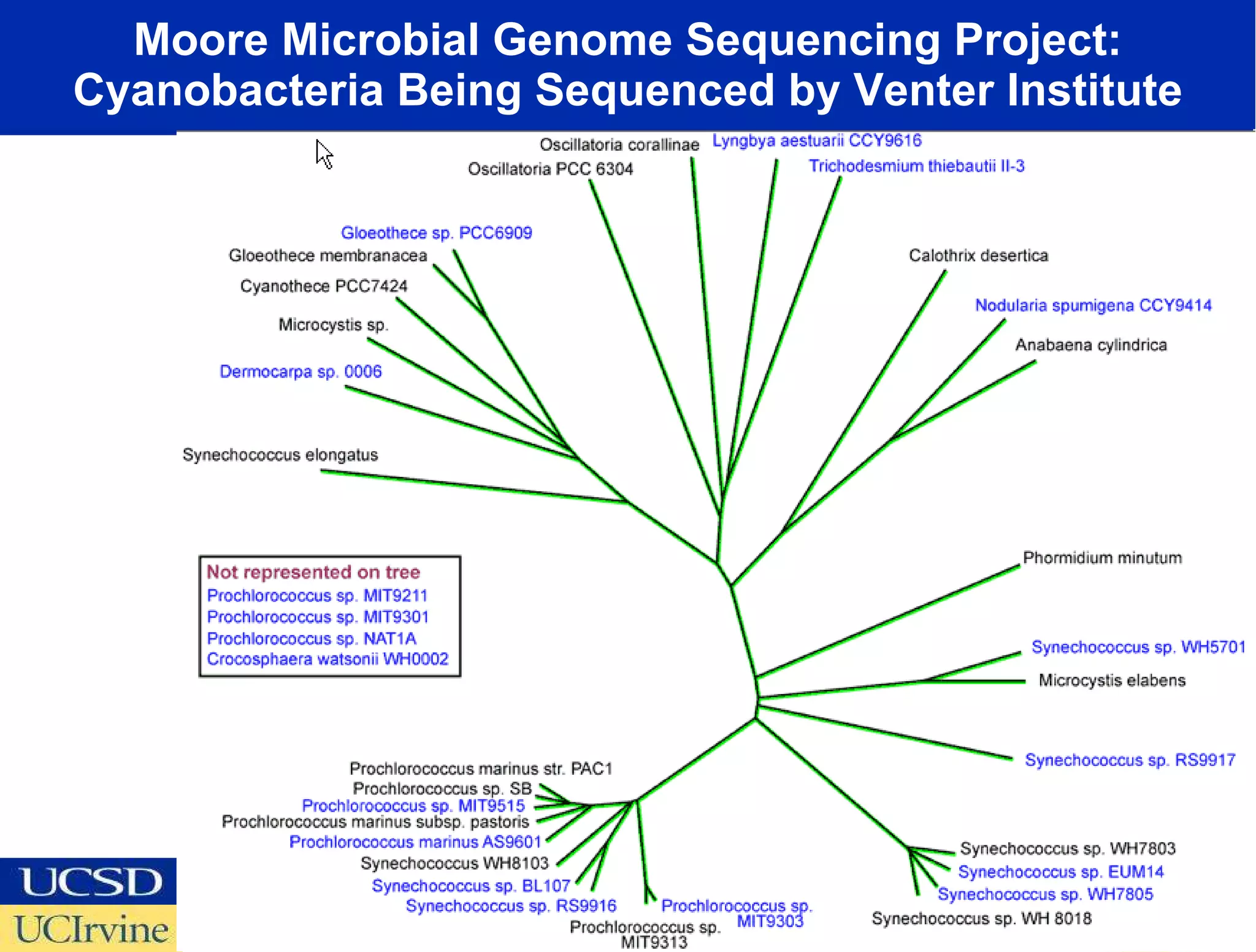
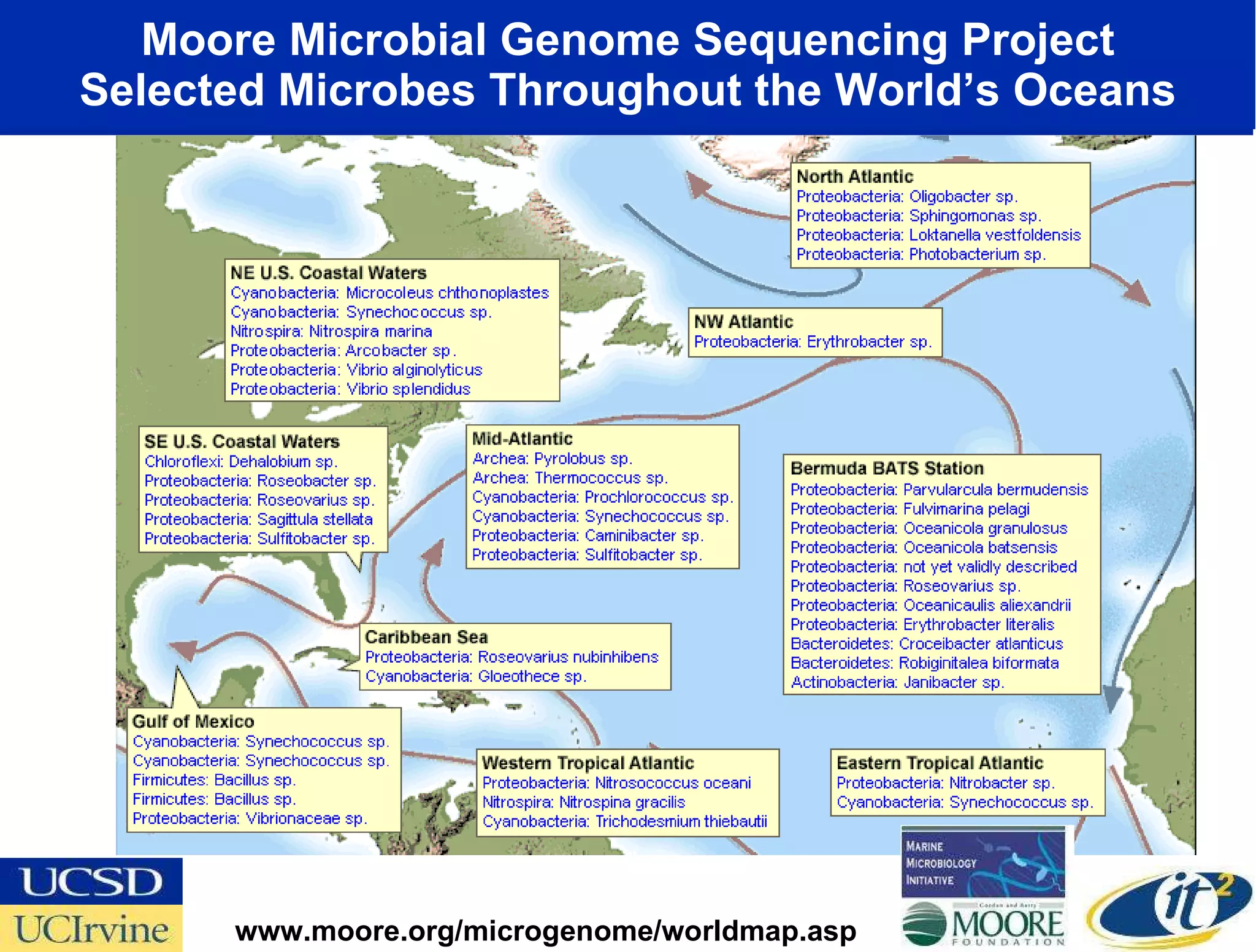
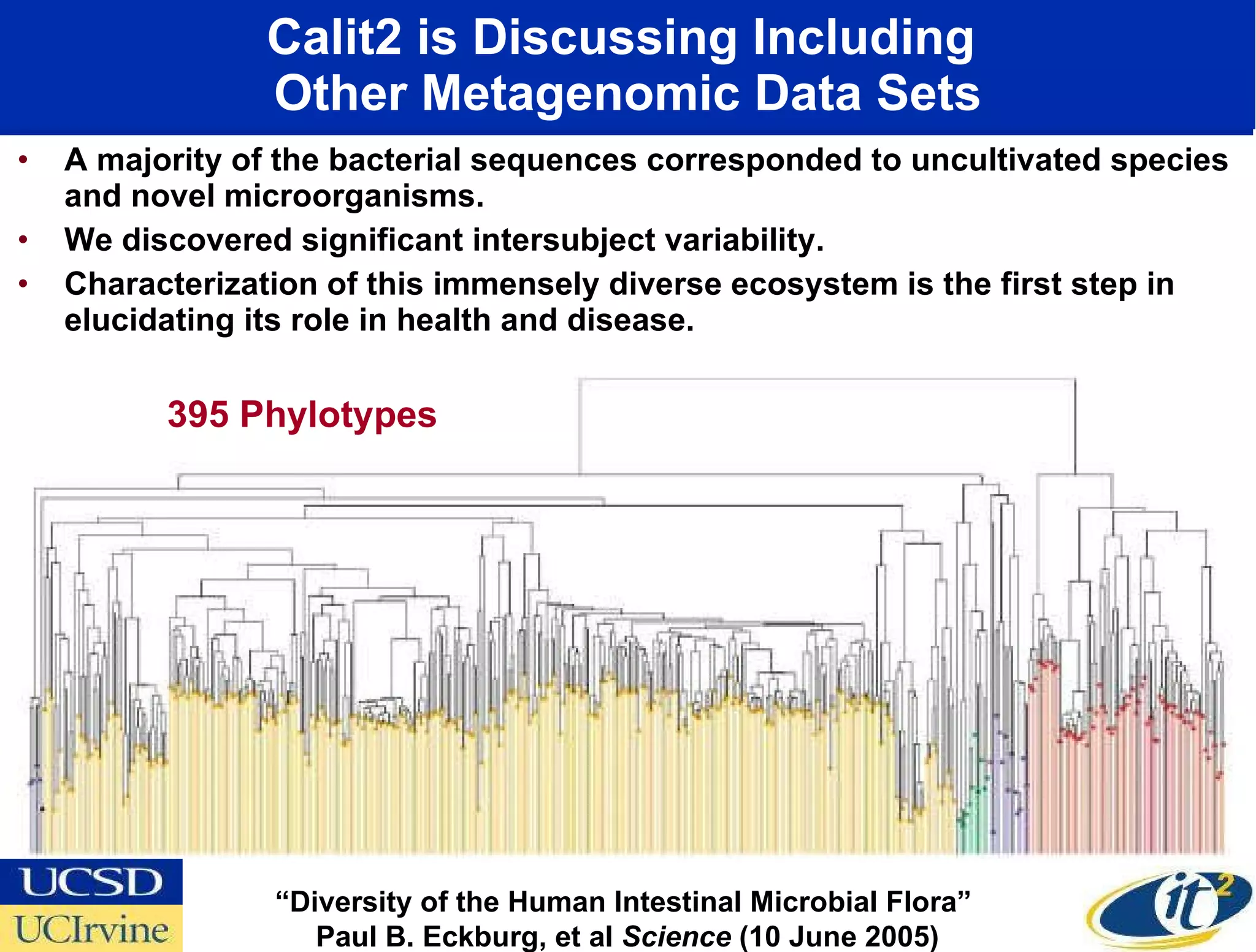
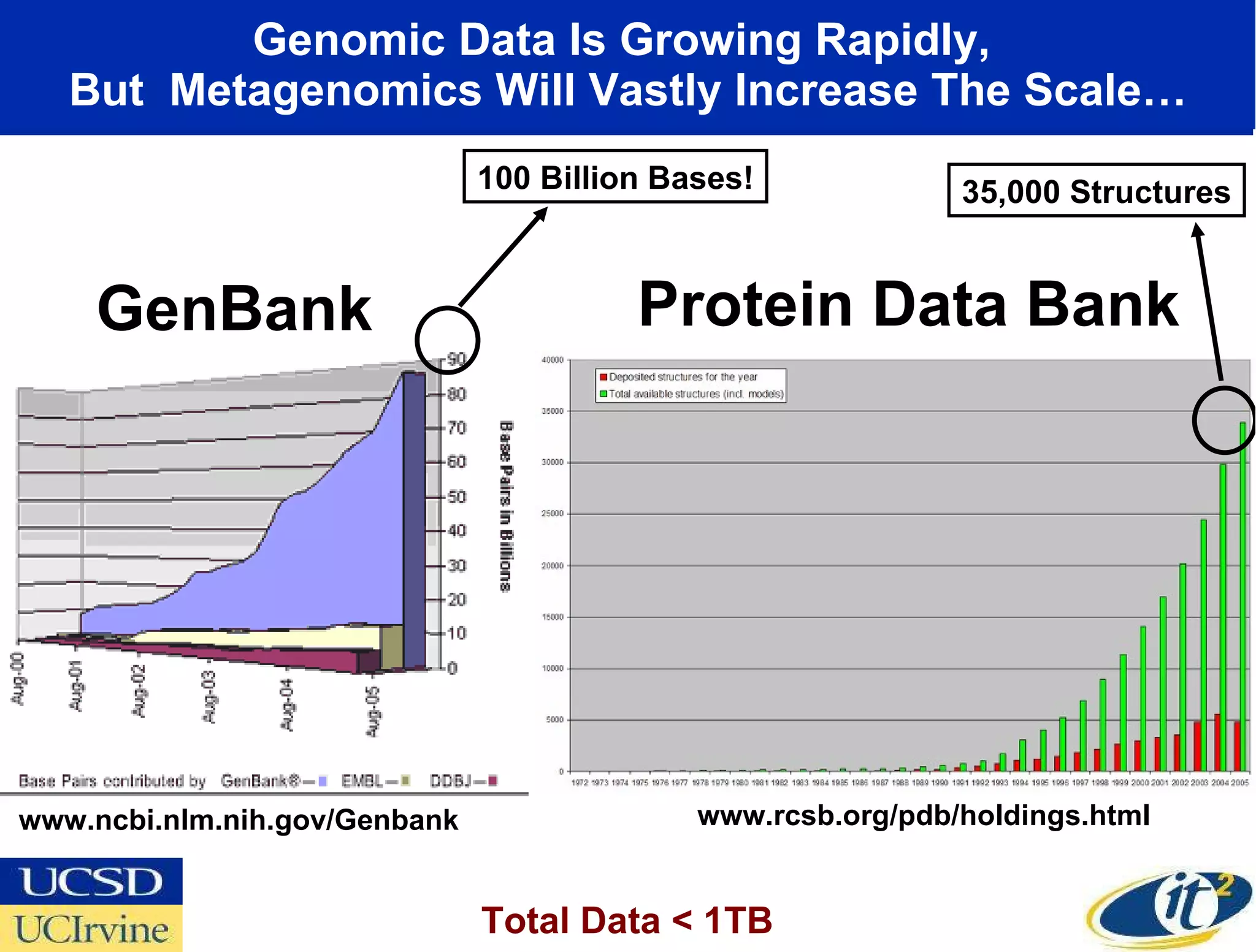
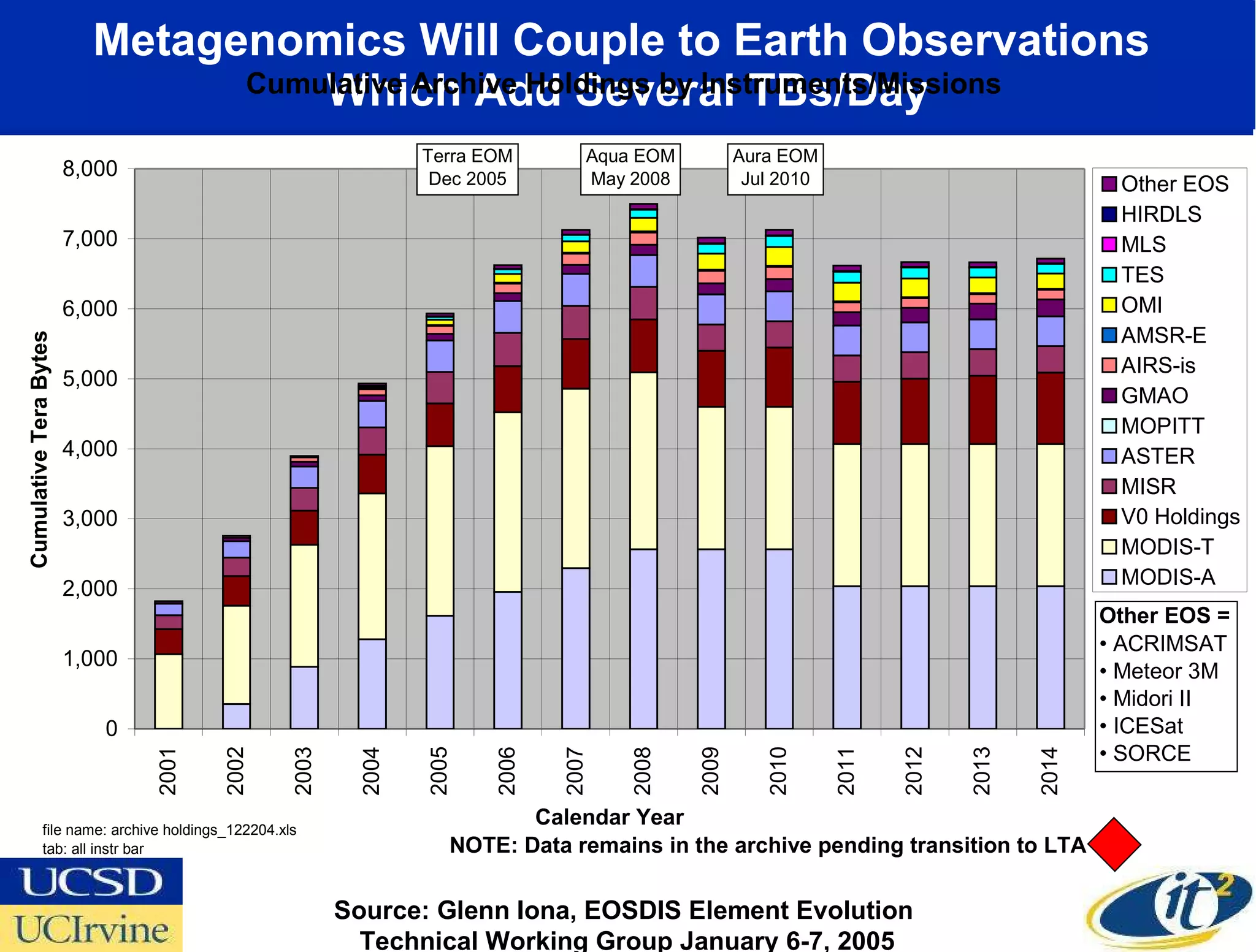
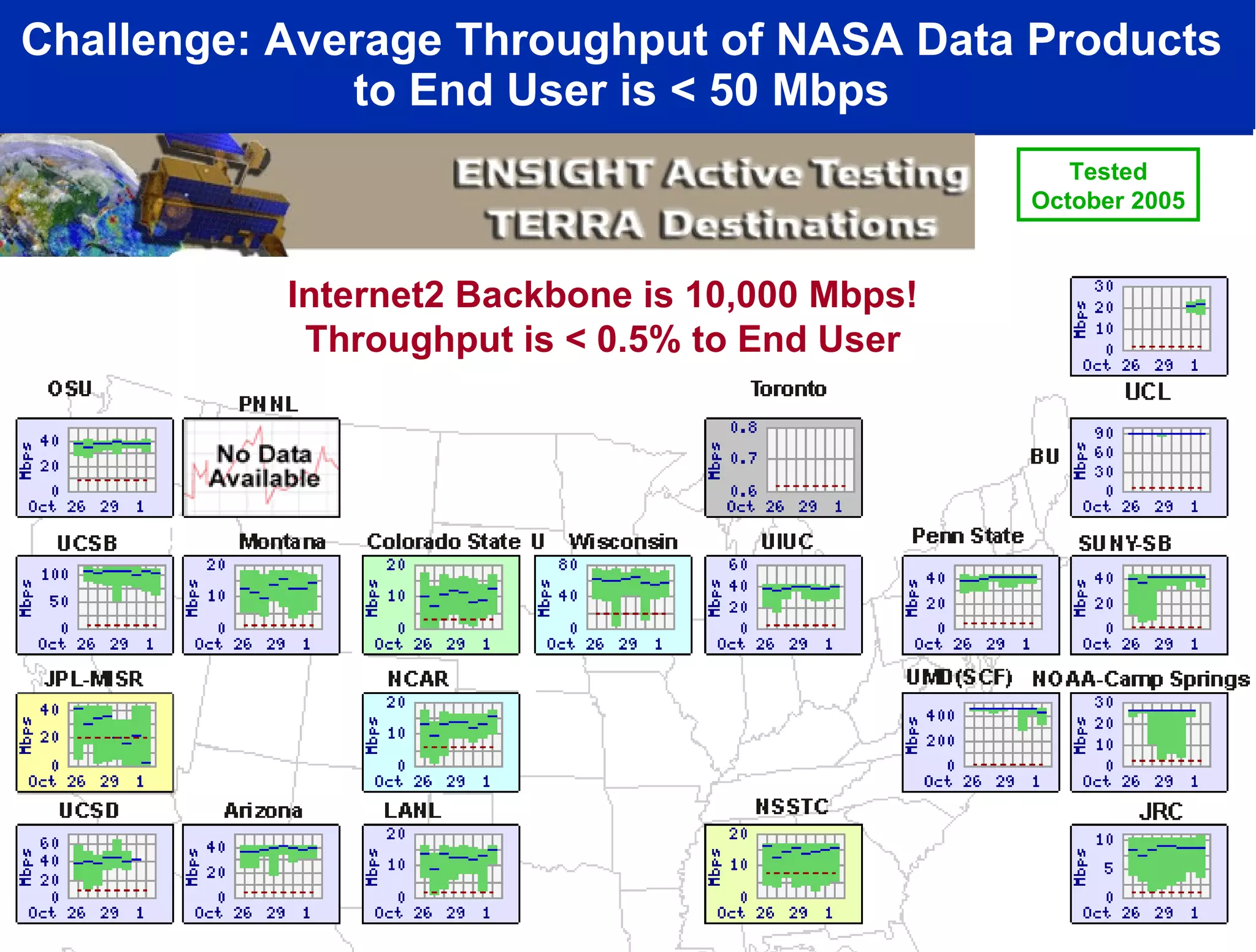
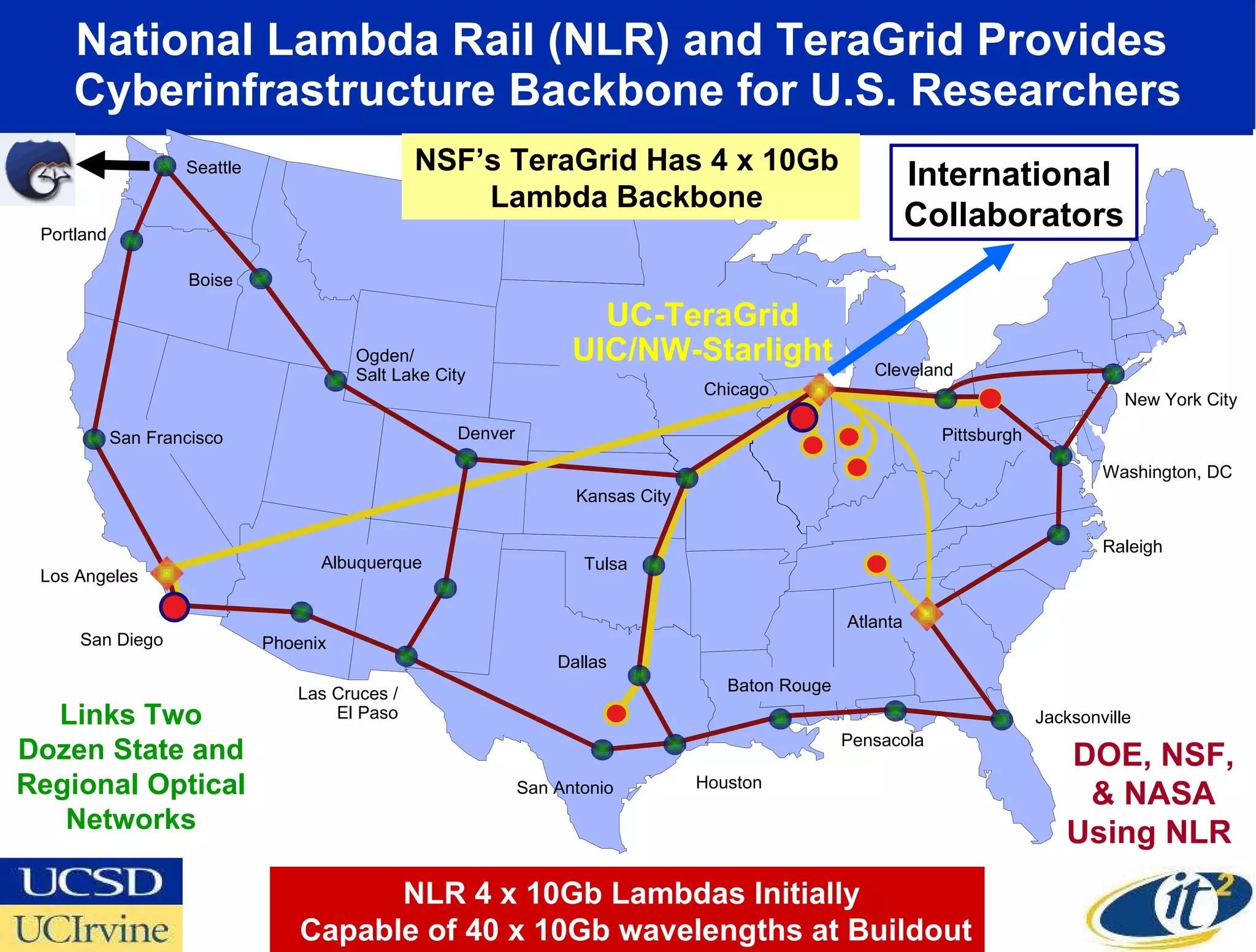
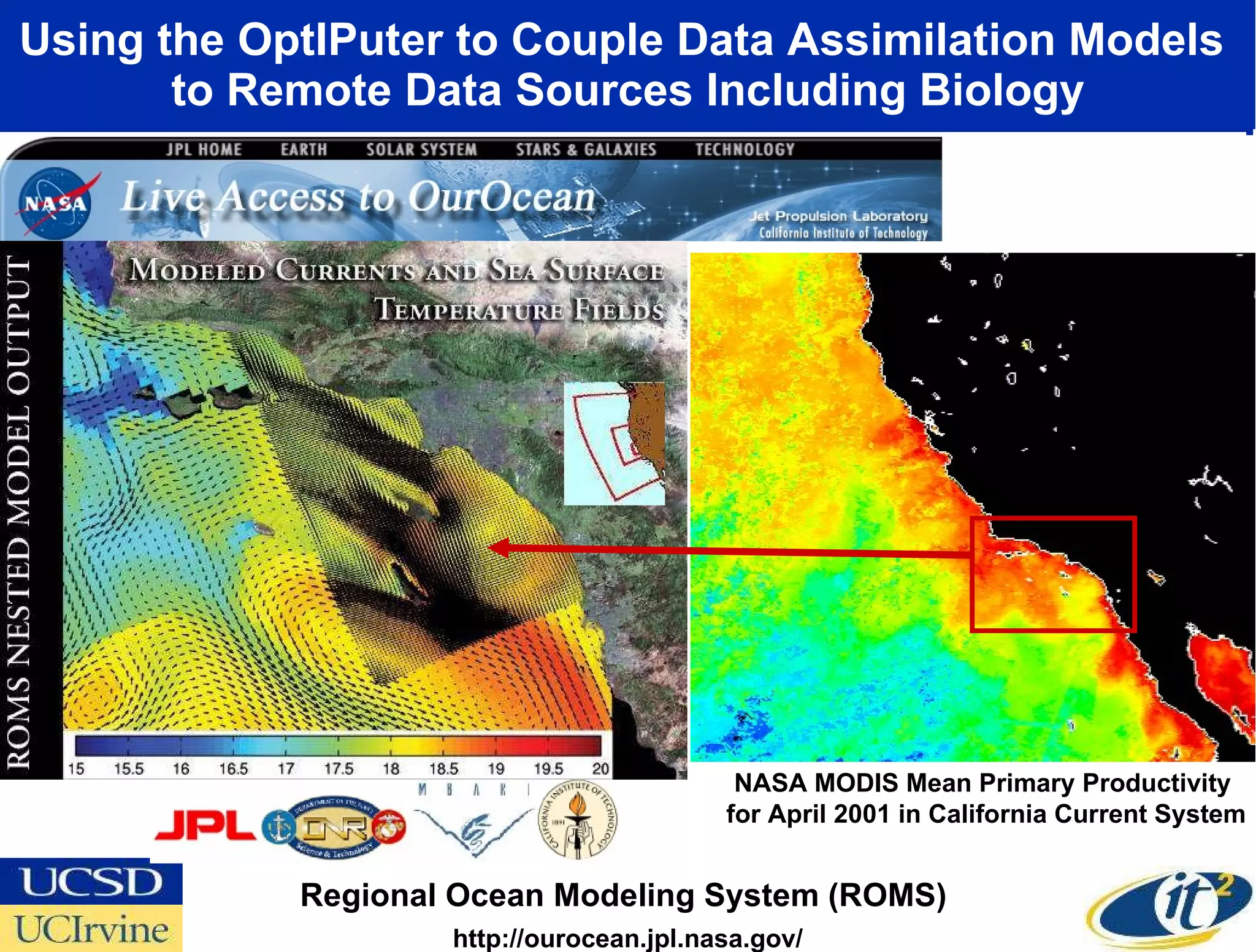
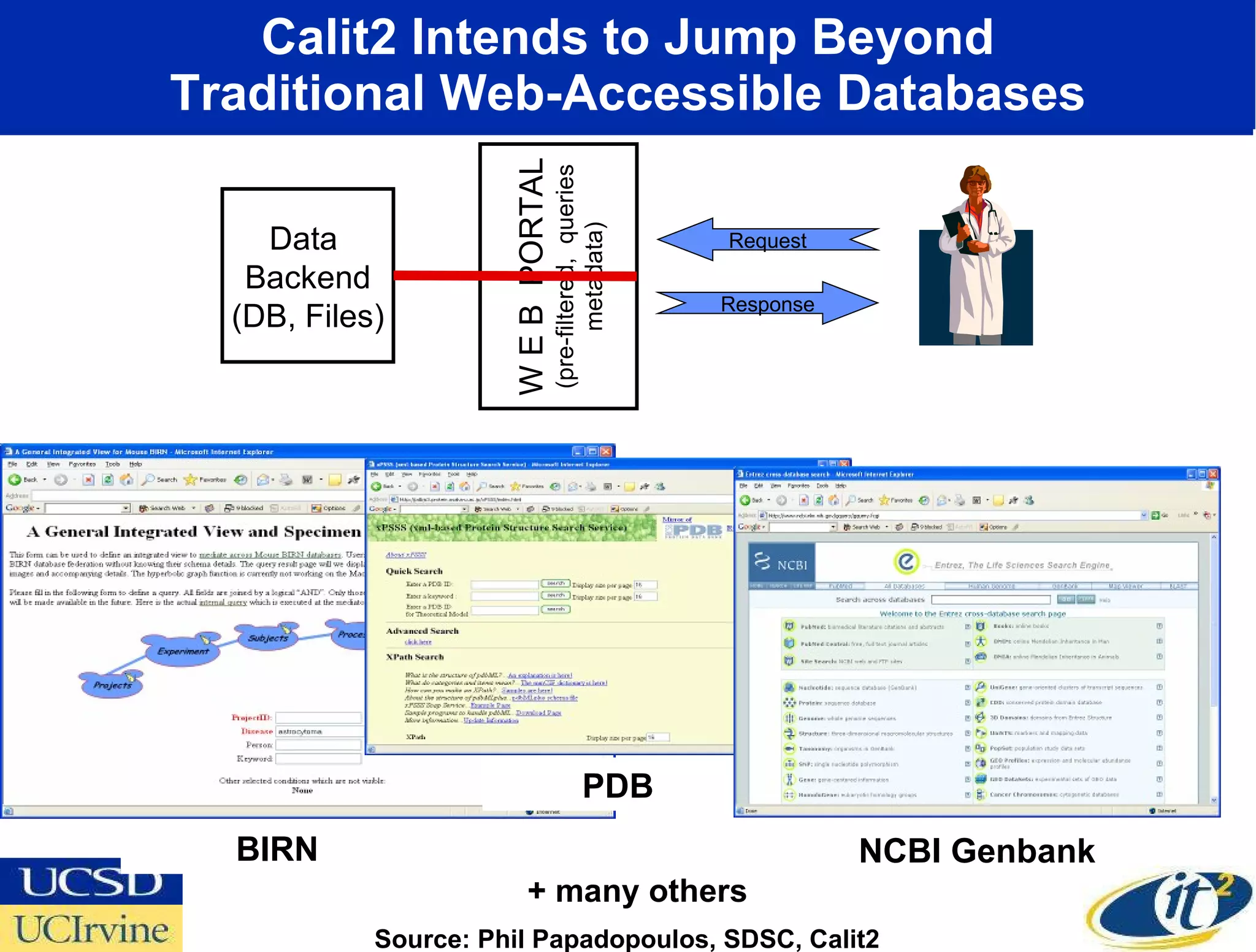
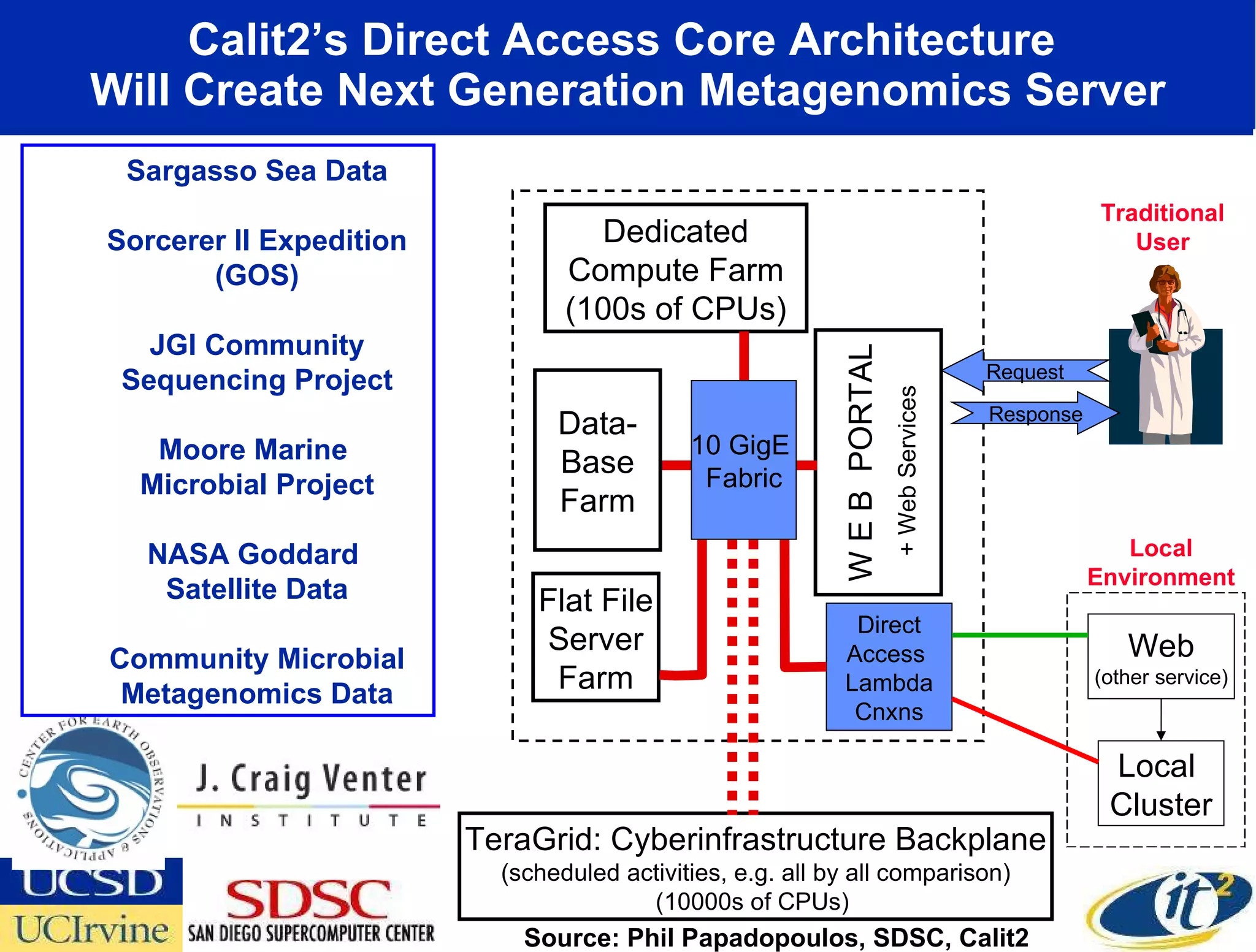
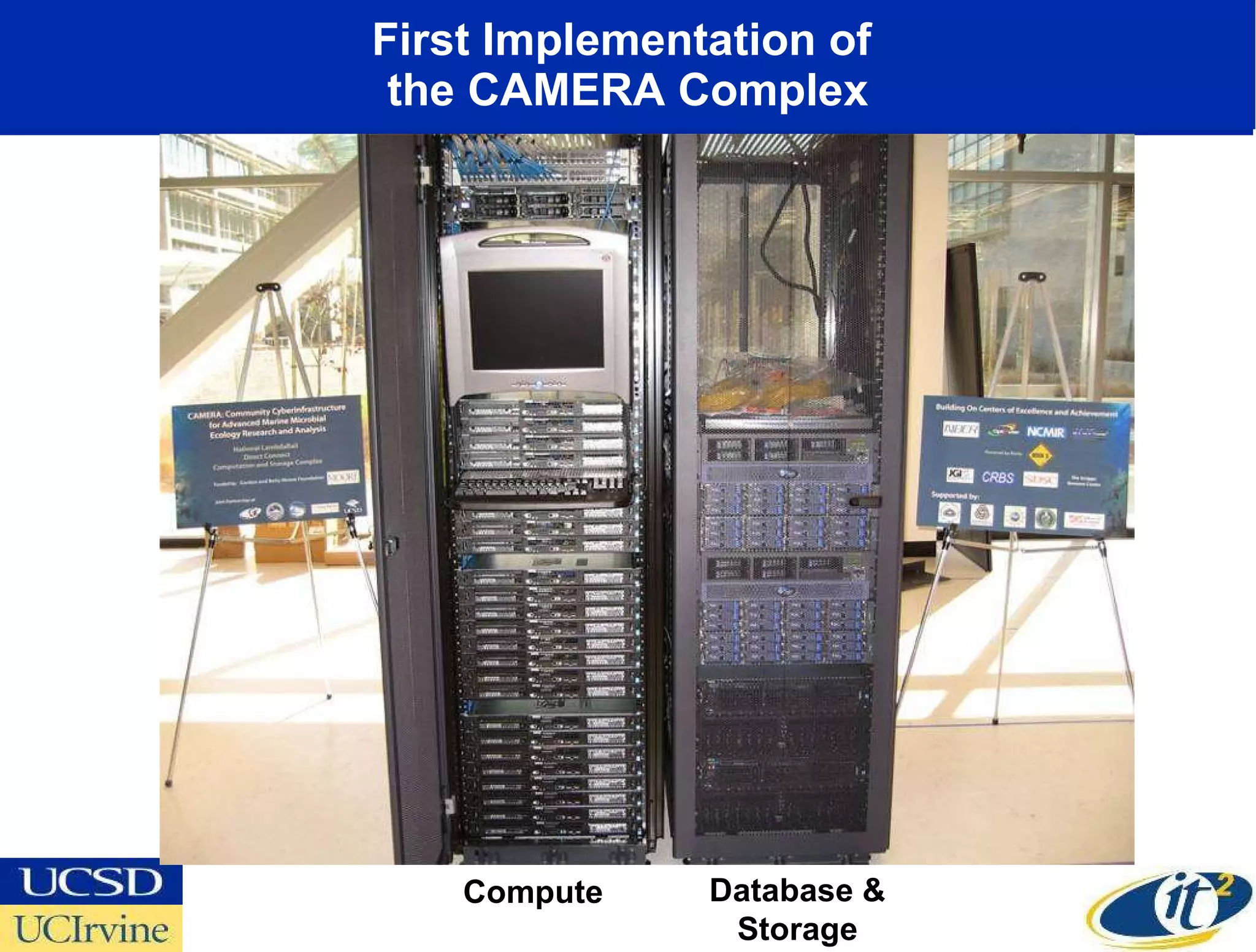
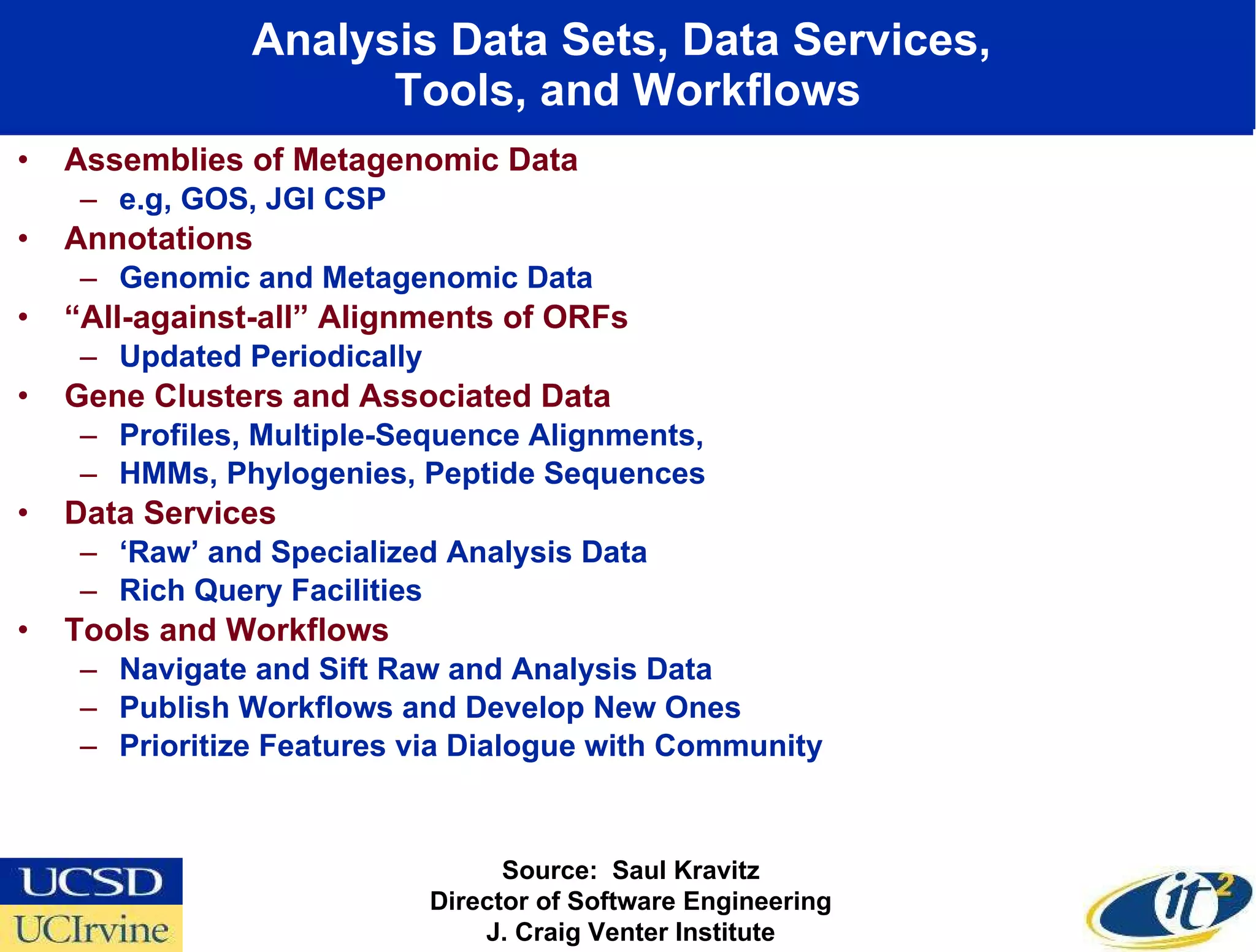
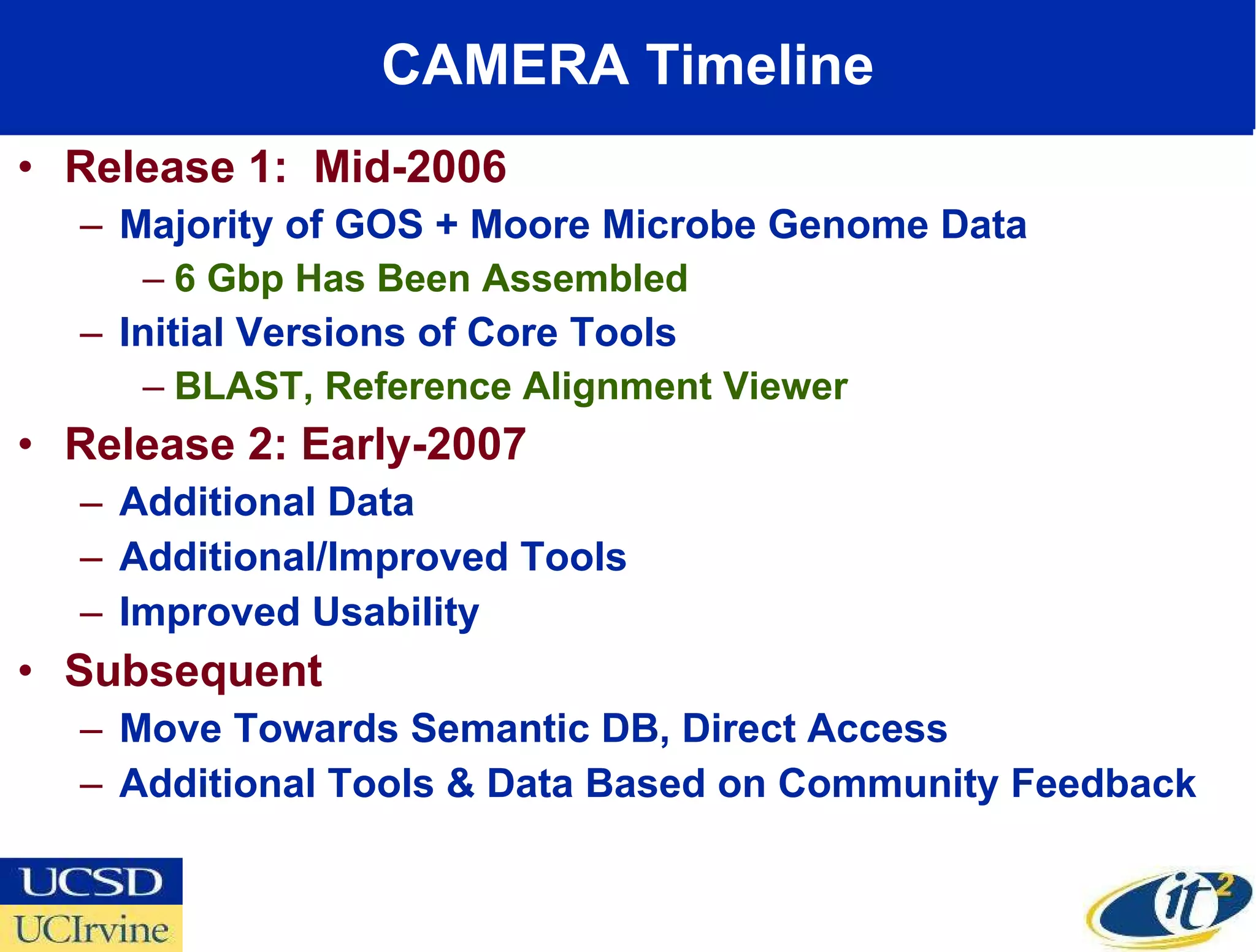
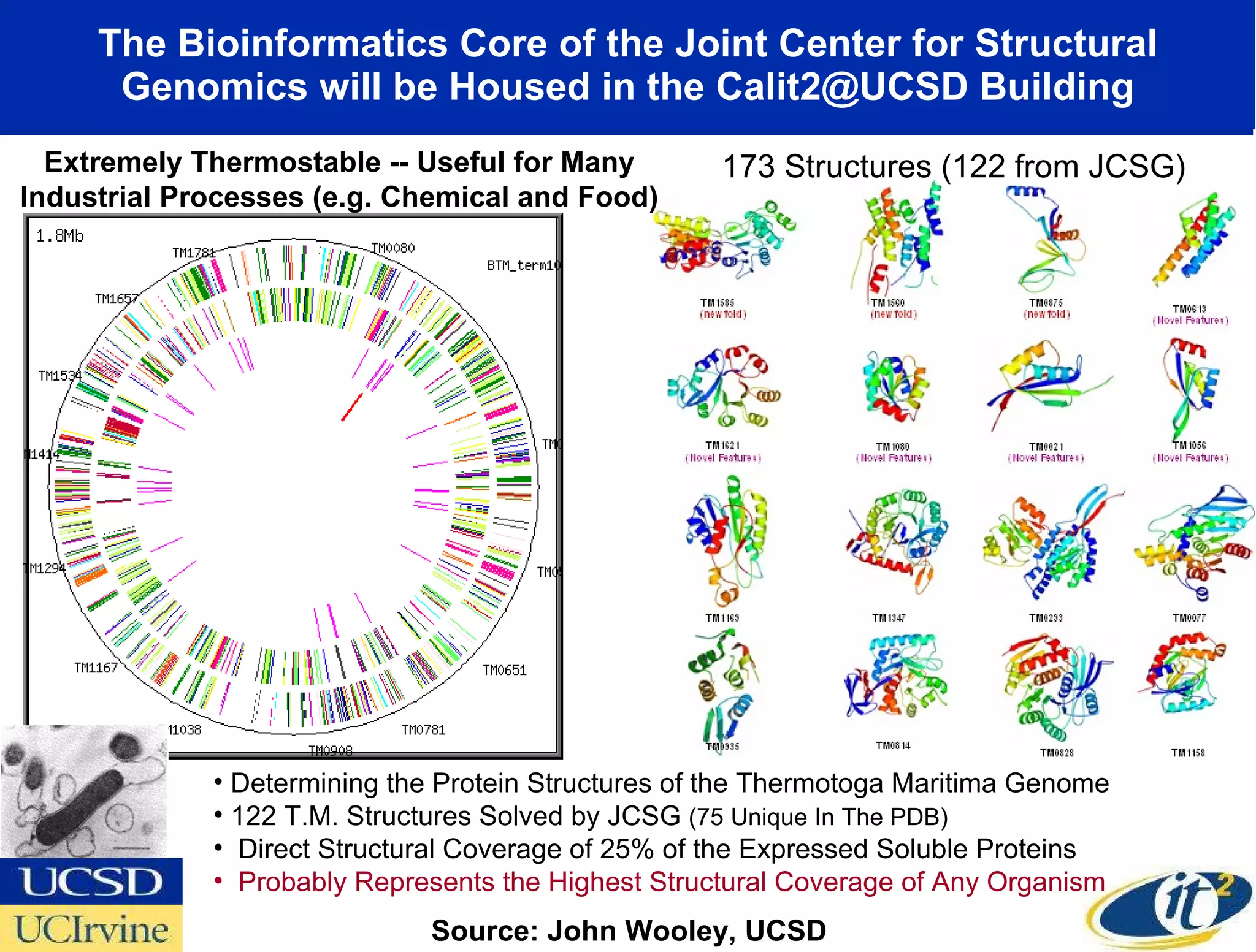
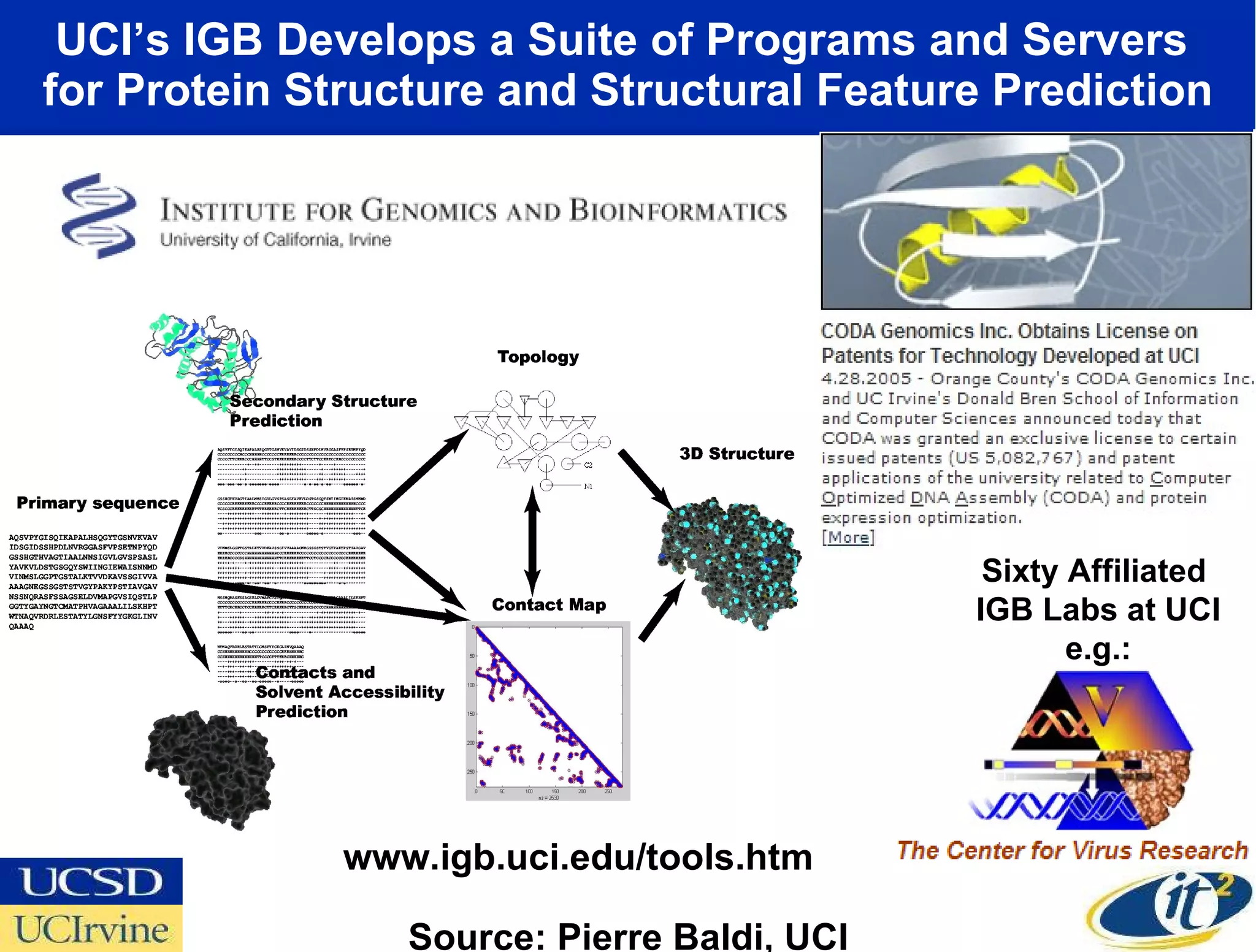
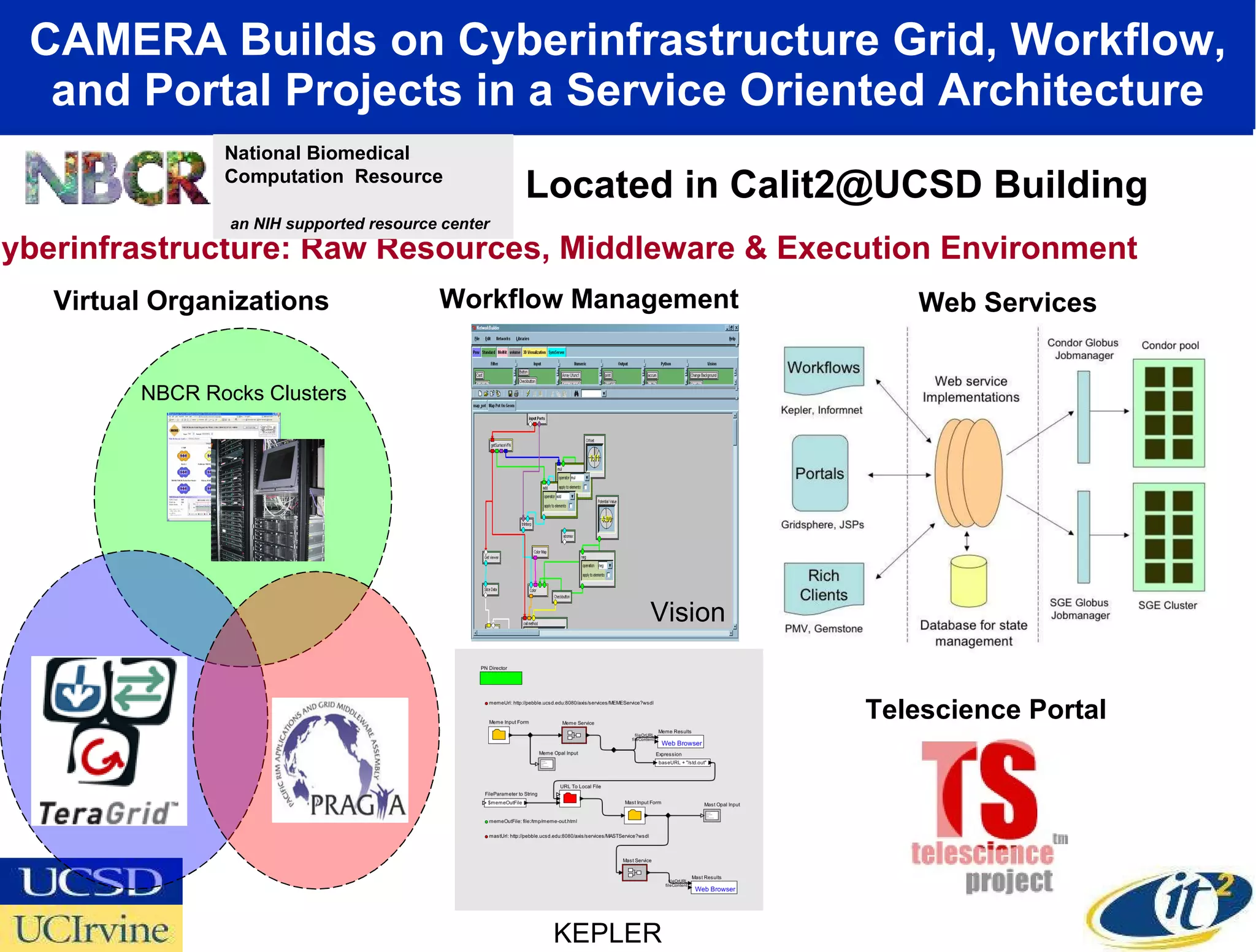
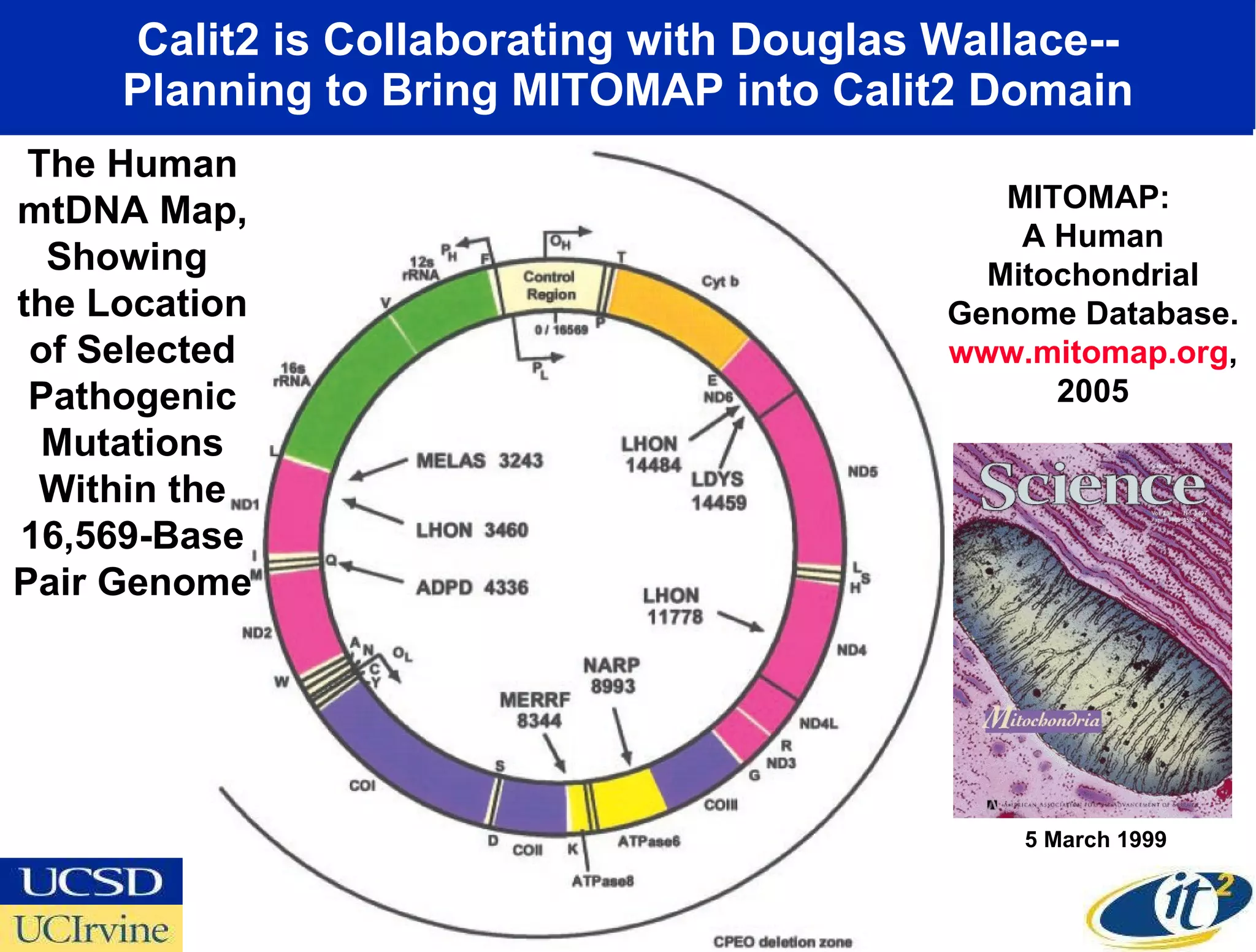
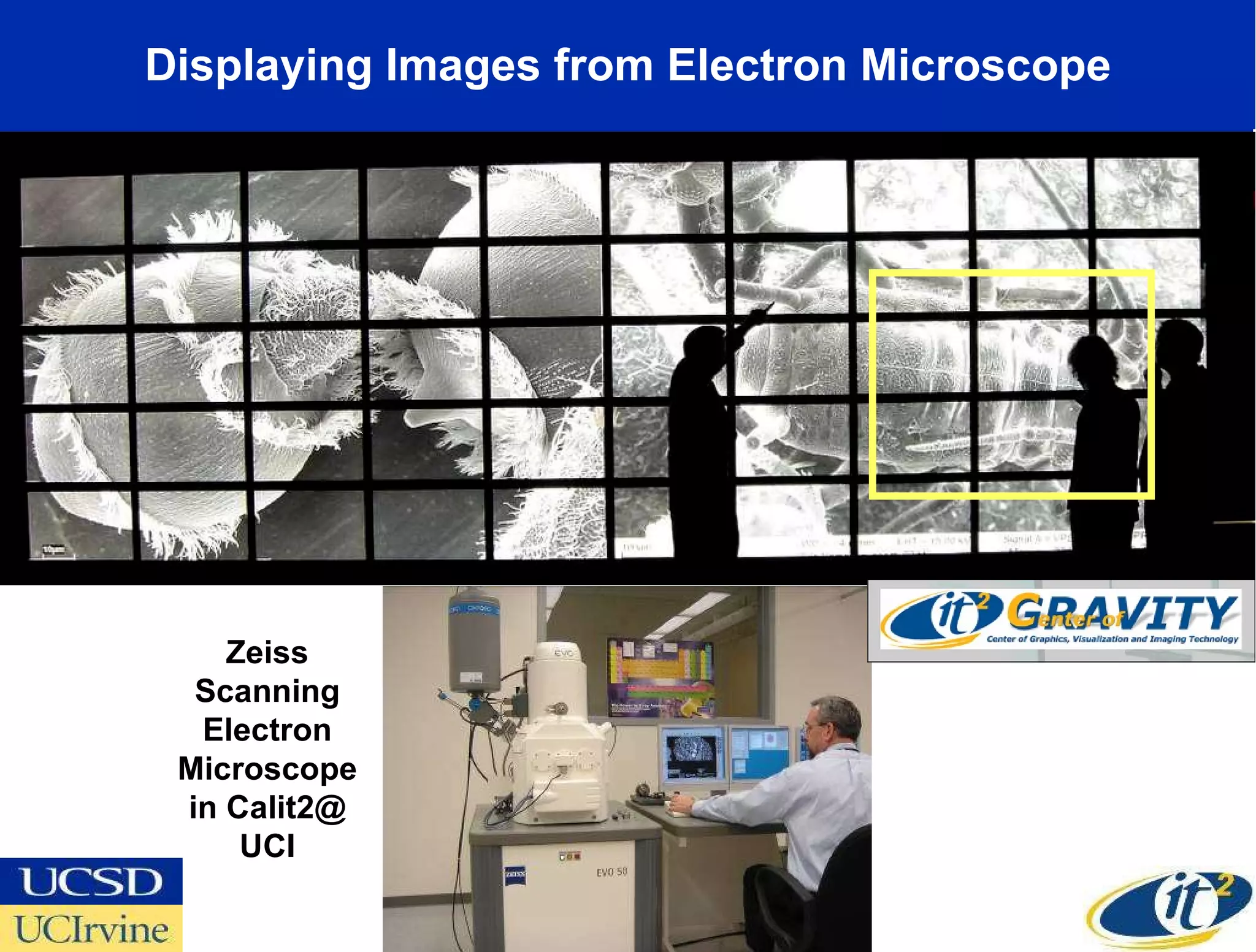
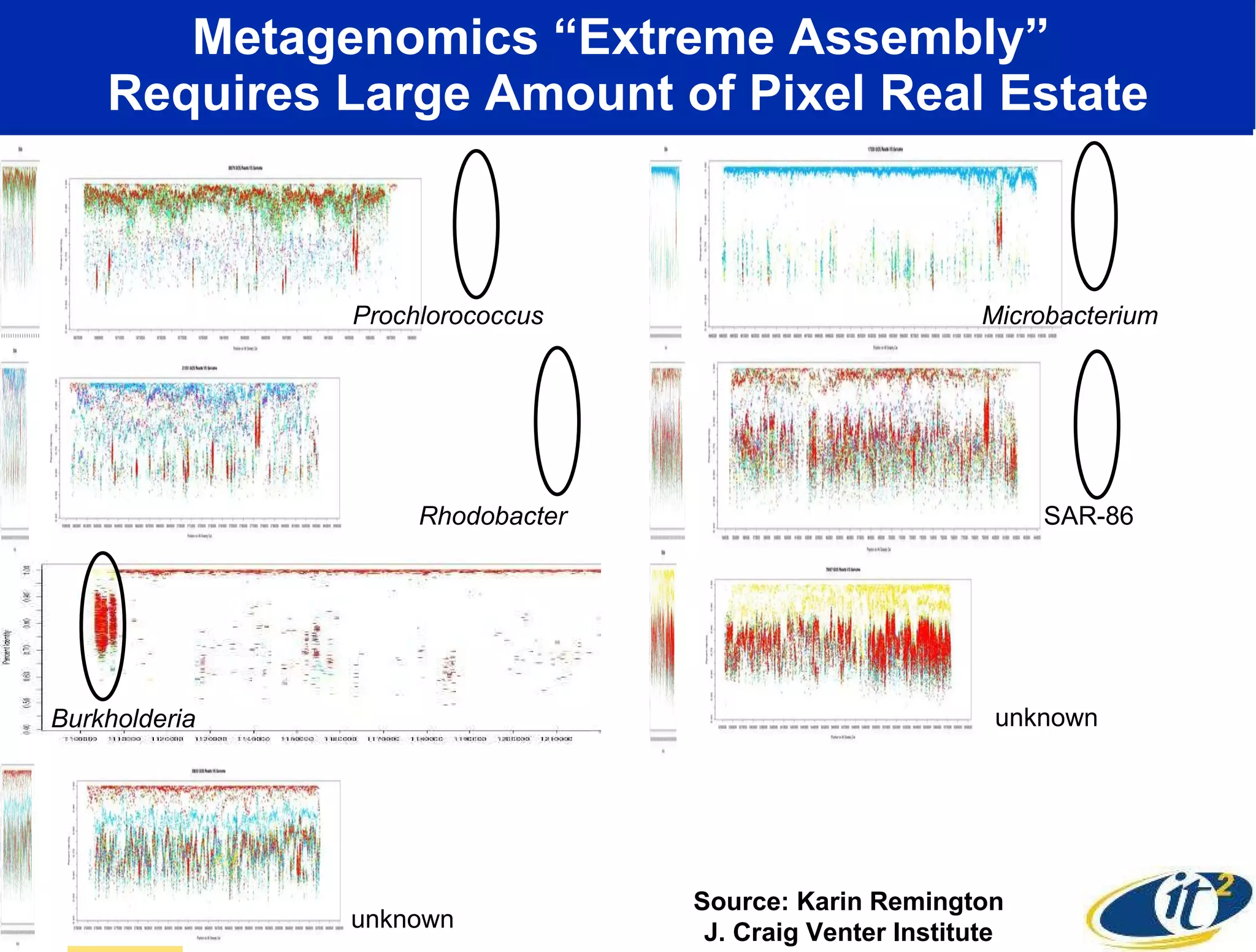
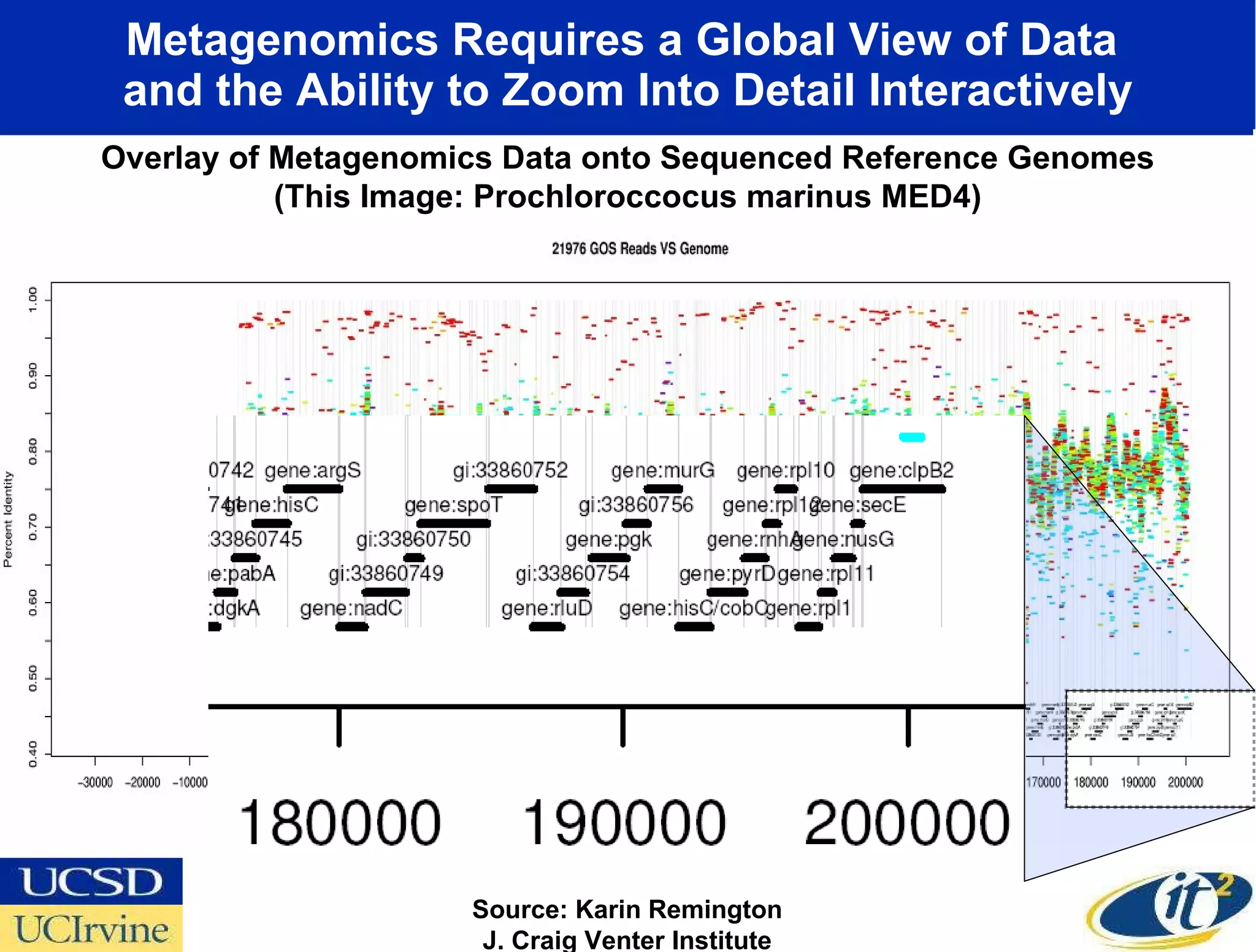
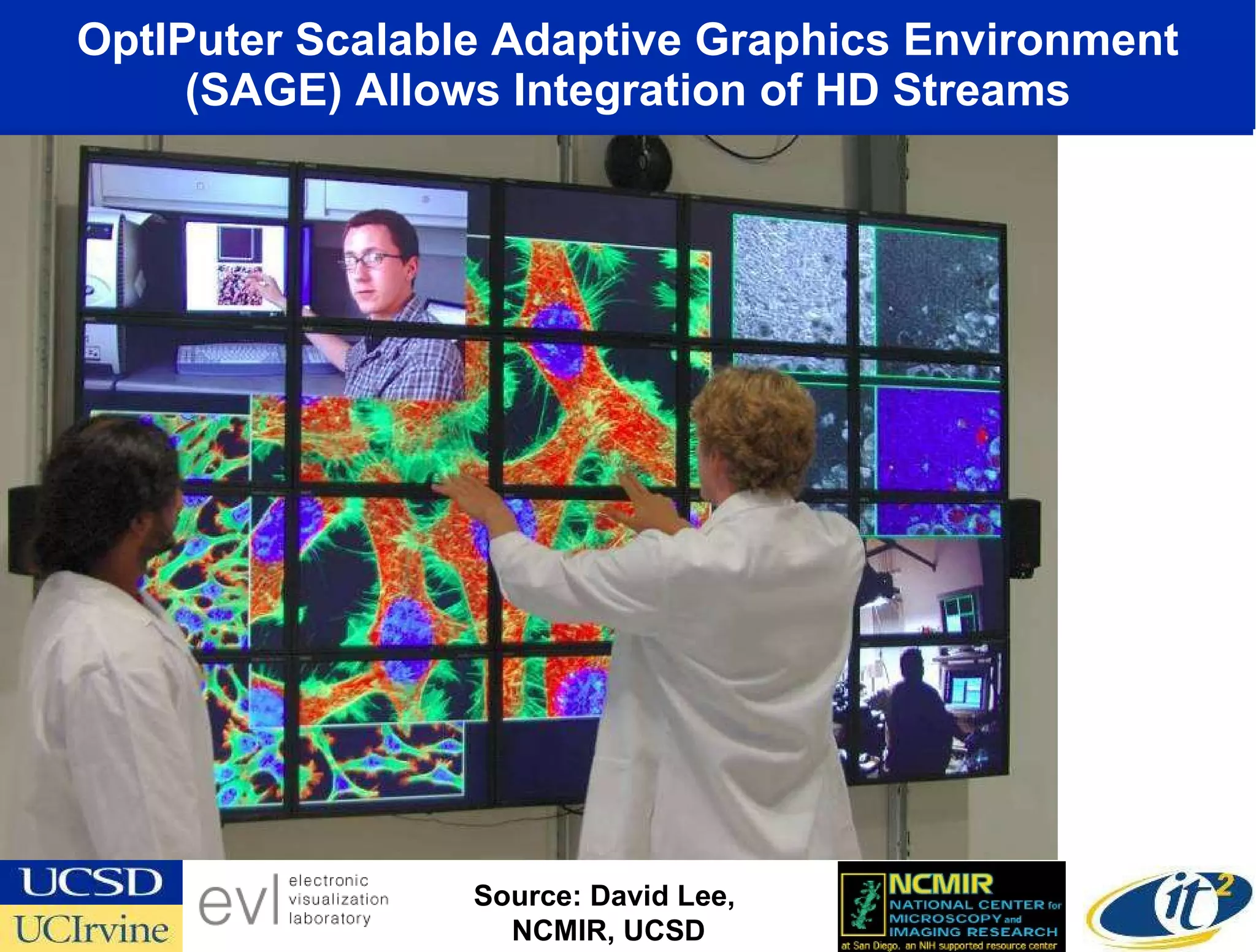
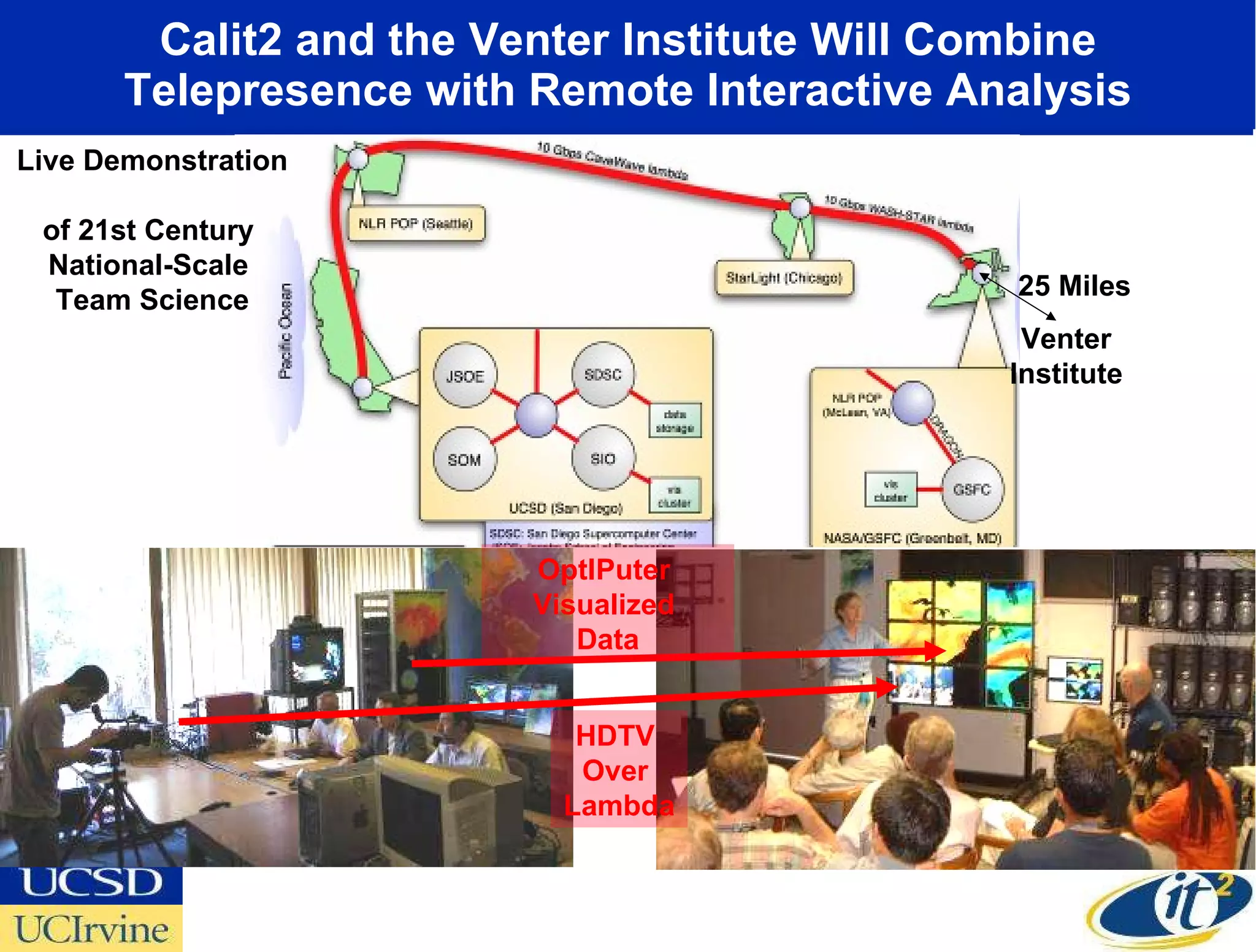
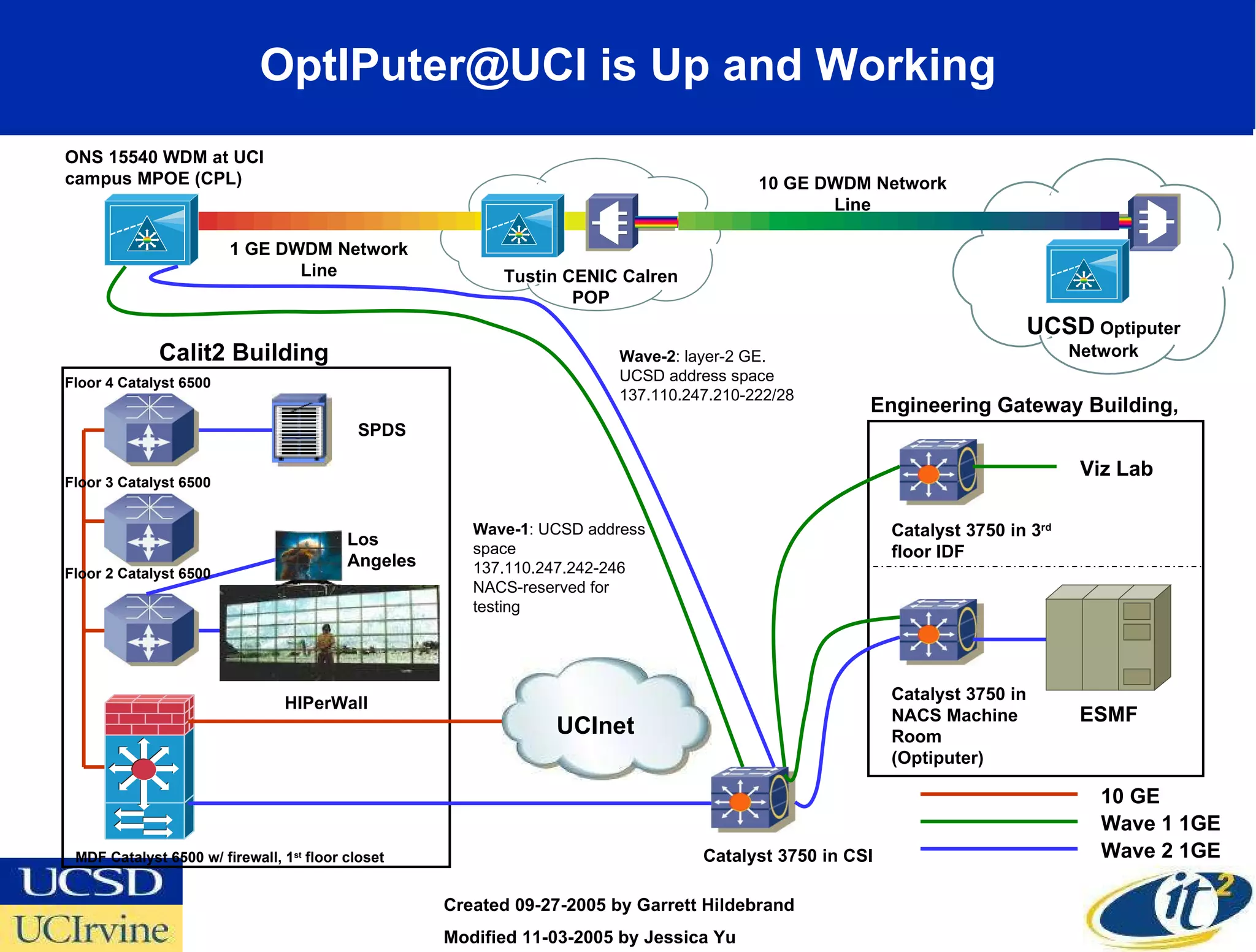
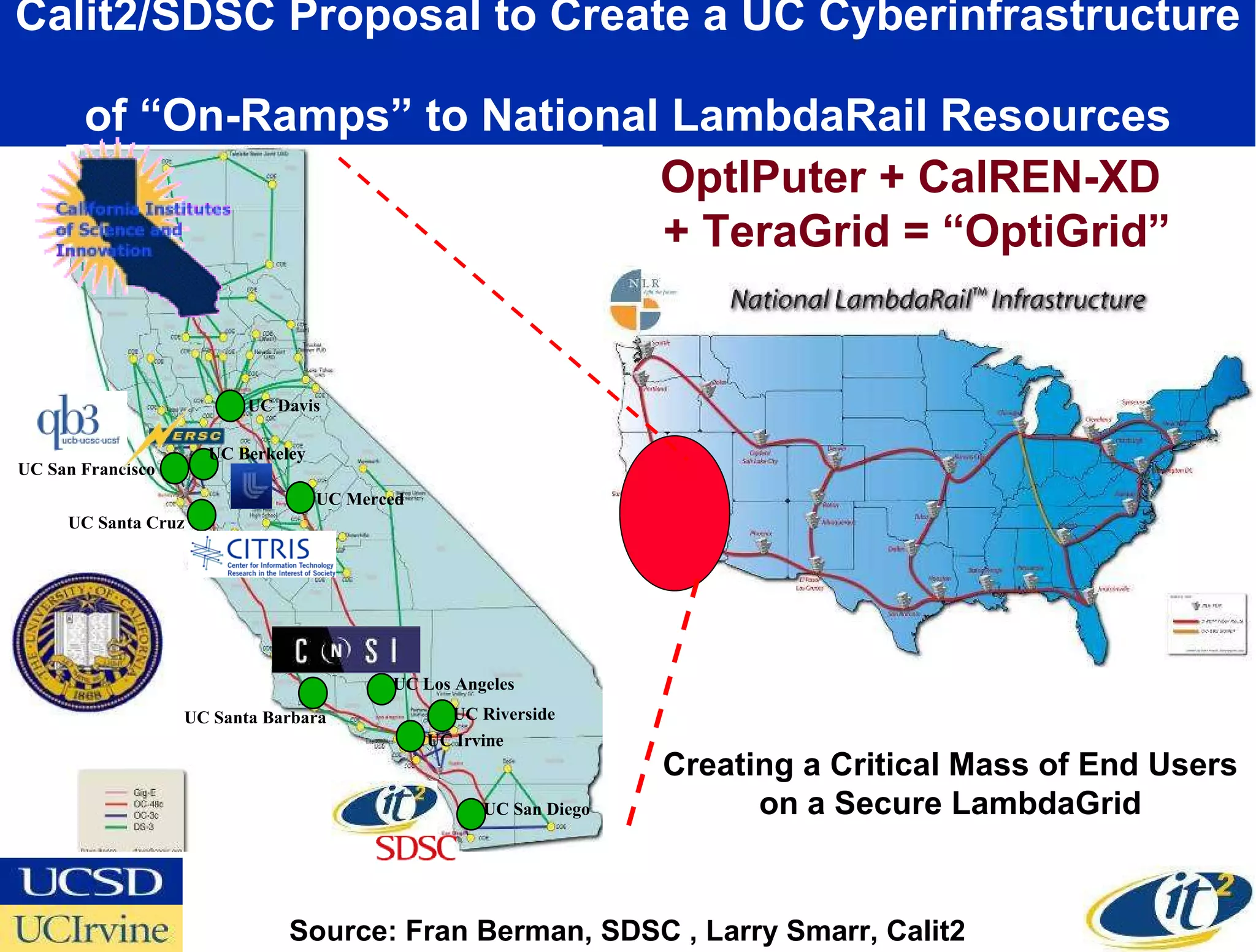
The document discusses the establishment of a computational resource and software tools by calit2, the venter institute, and ucsd's center for earth observations to analyze the genetic code of marine microbial communities, funded by a $24.5 million grant from the moore foundation. This initiative includes the creation of the community cyberinfrastructure for advanced marine microbial ecology research and analysis (camera), which will enhance metagenomic research and expand the genomic database significantly. Furthermore, the project aims to bridge various scientific fields by integrating ecological genomic data and leveraging high-throughput sequencing technologies.