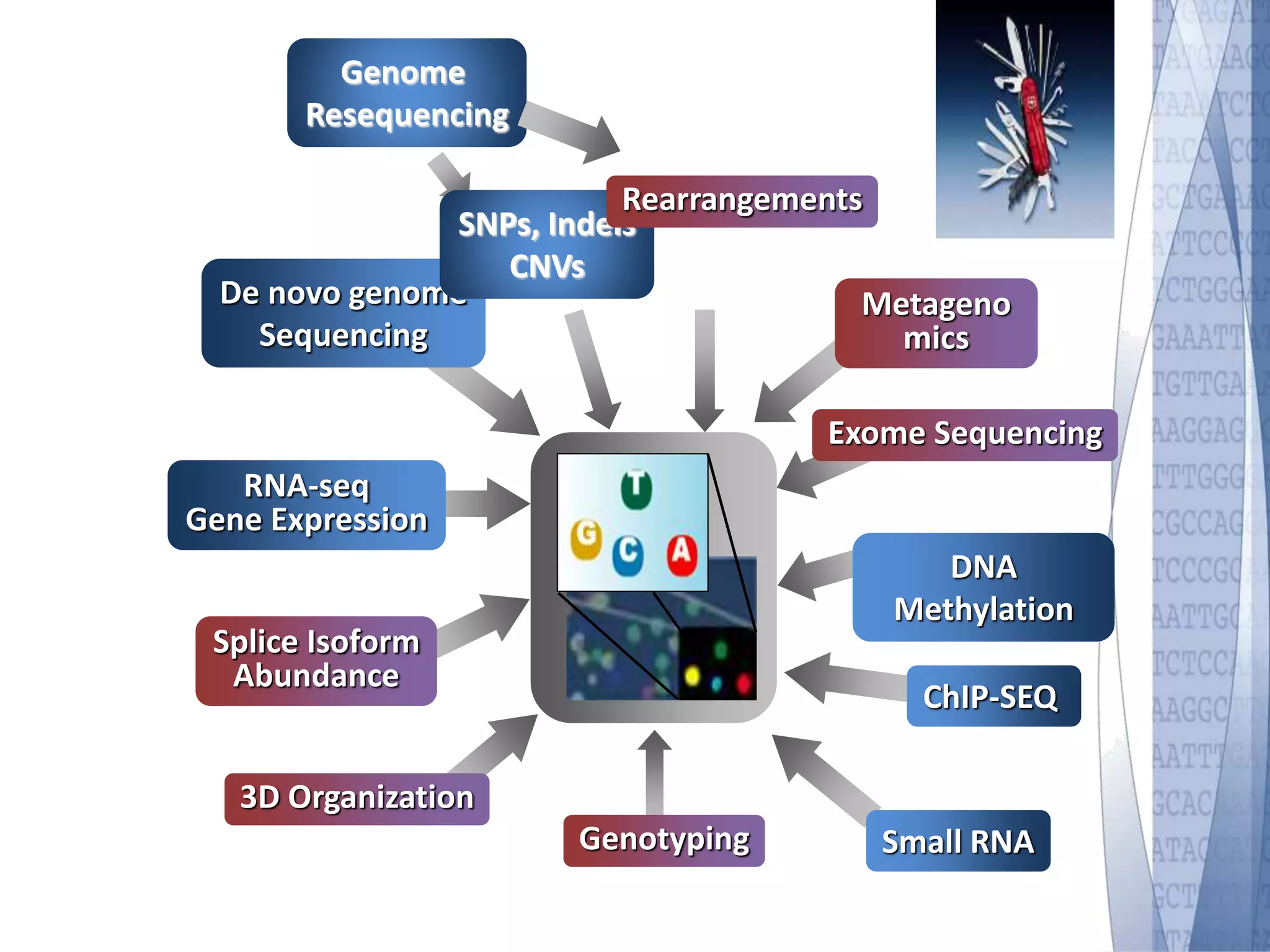
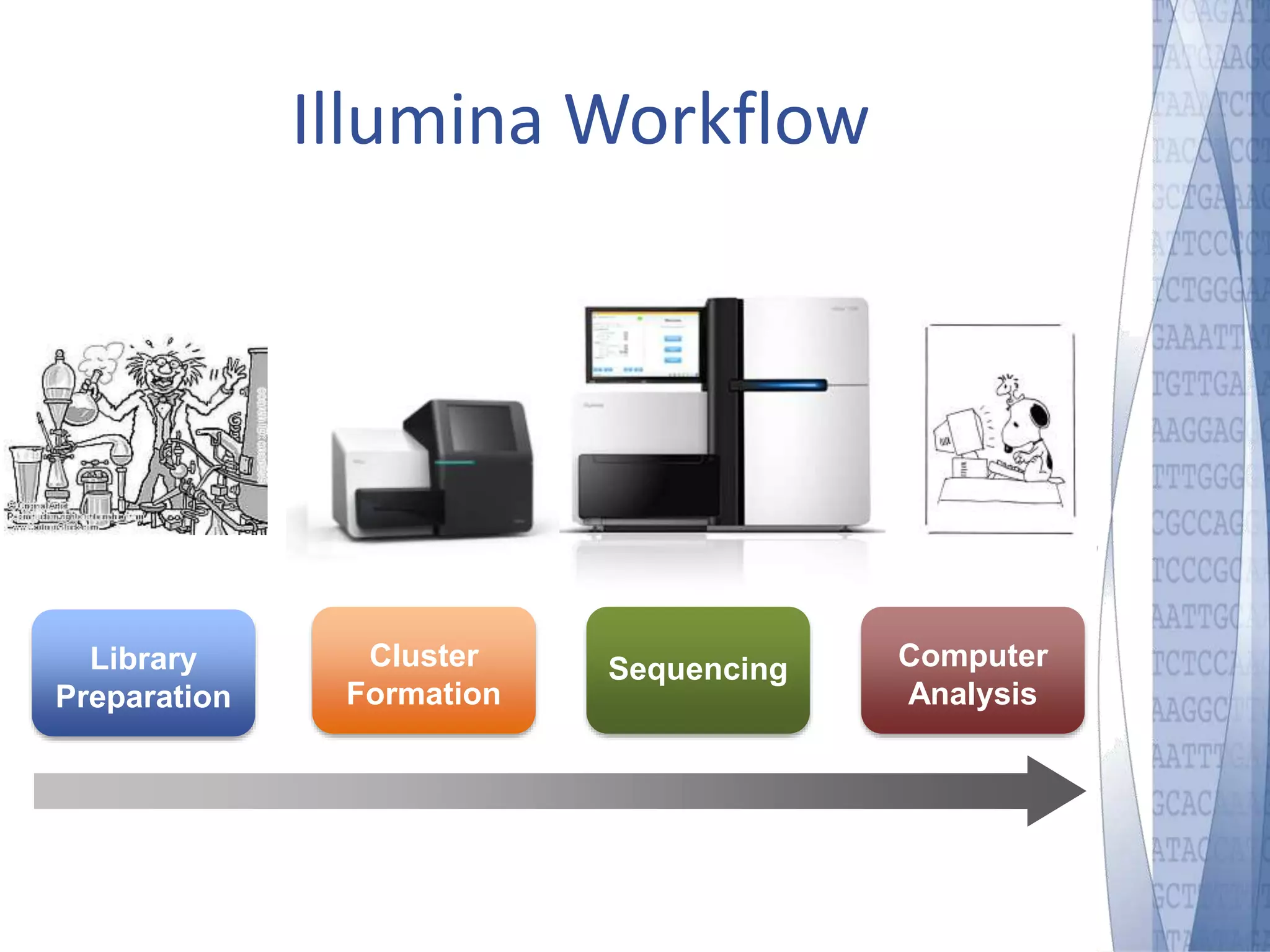
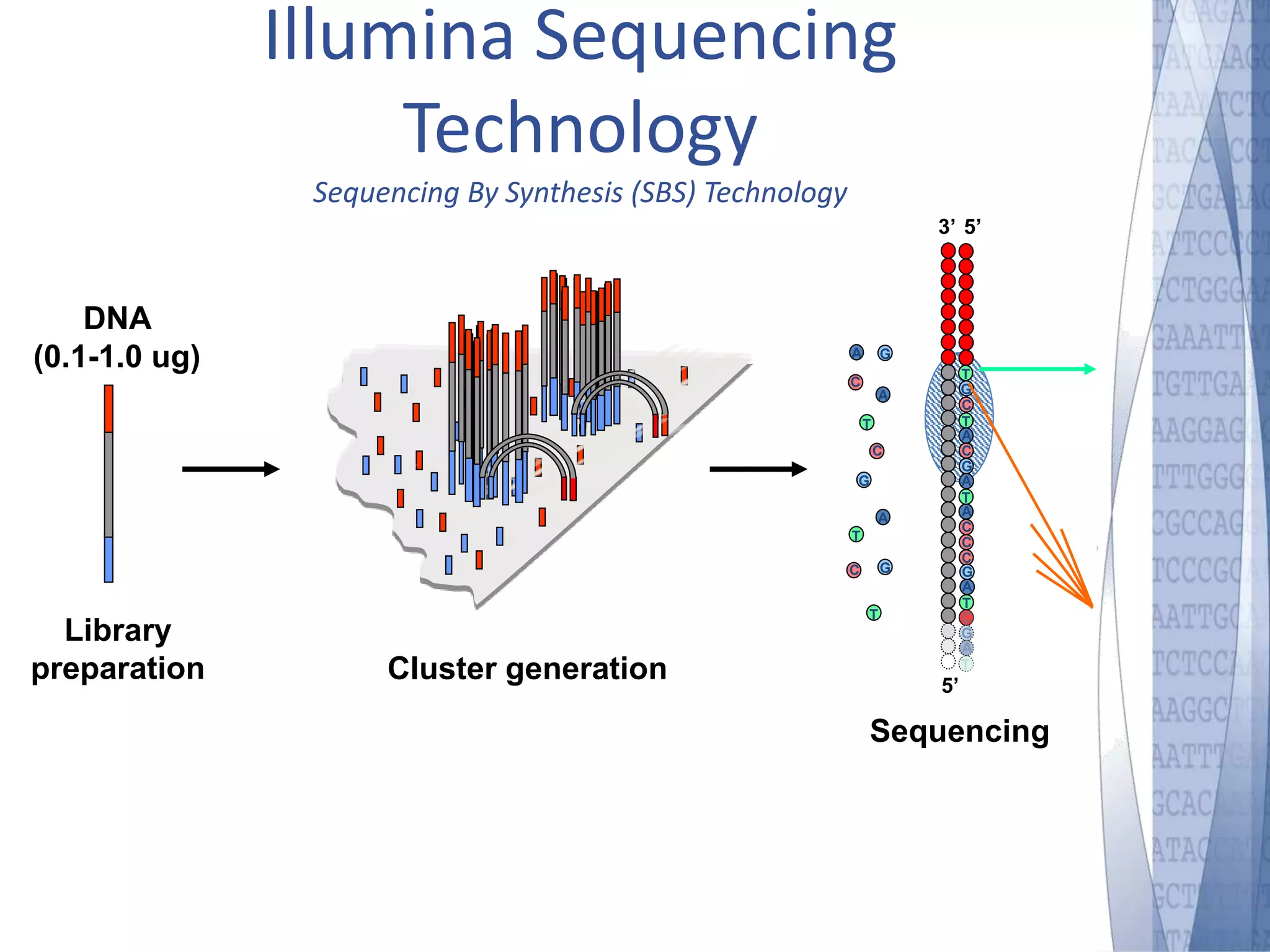
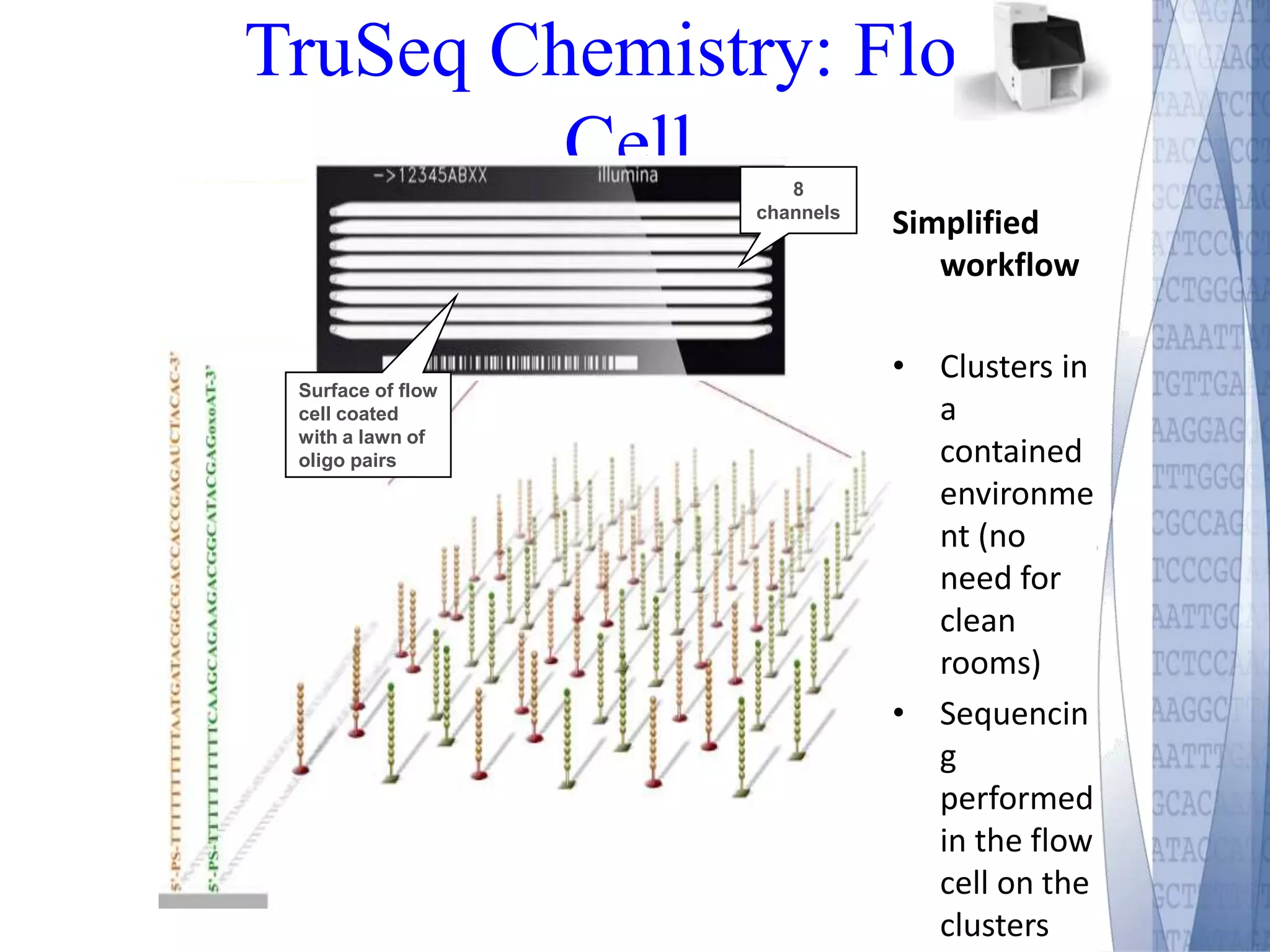
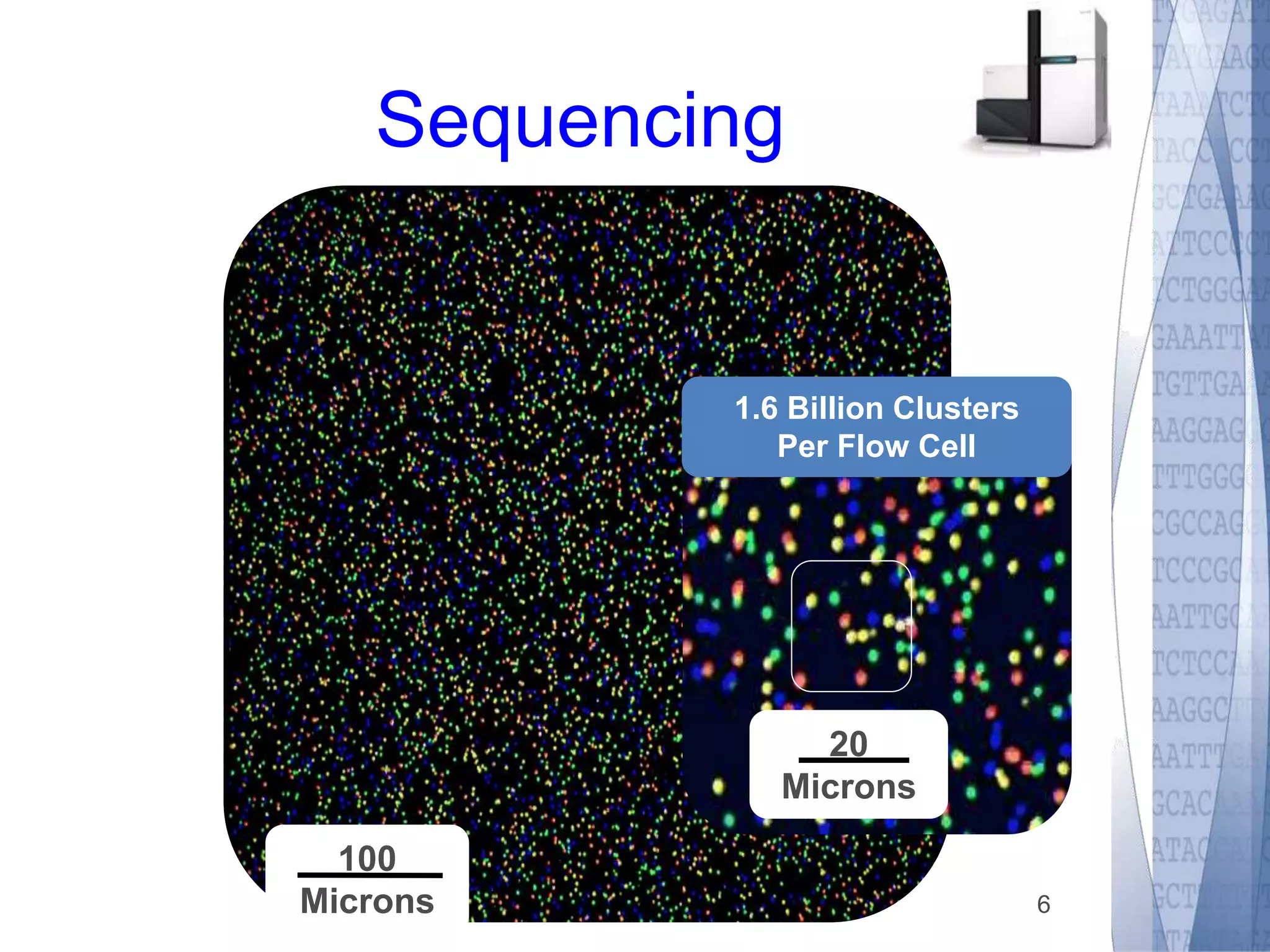
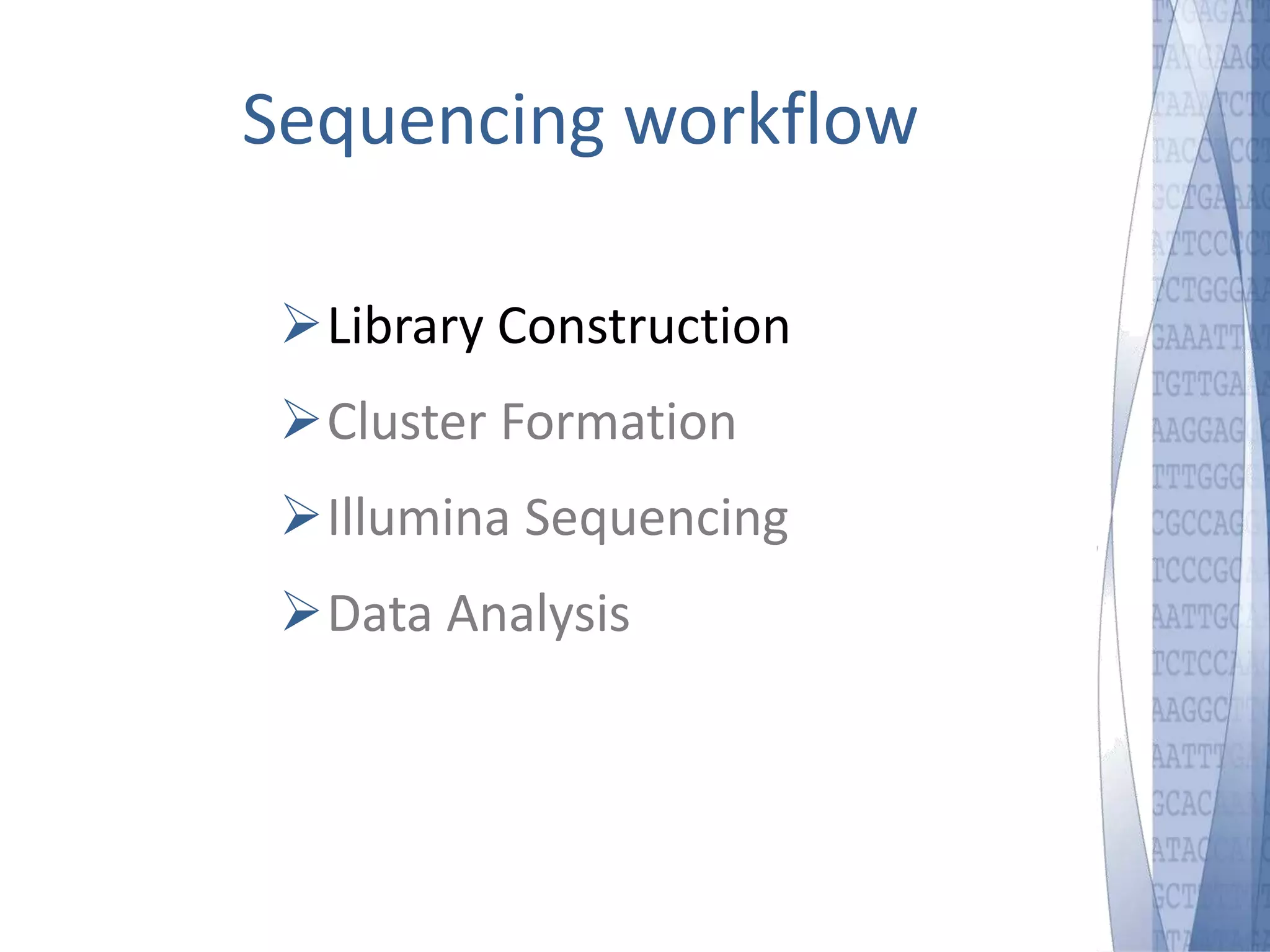
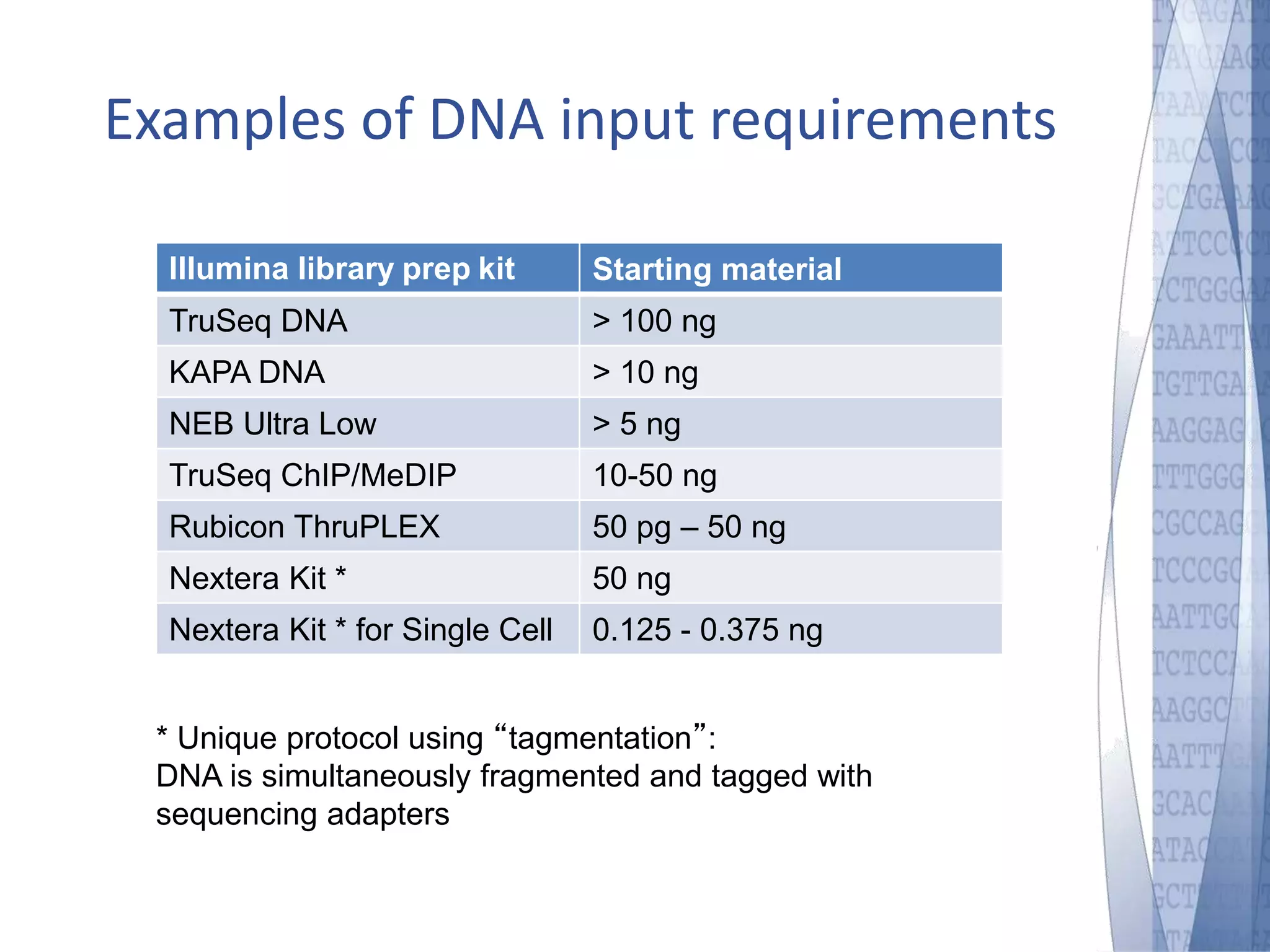
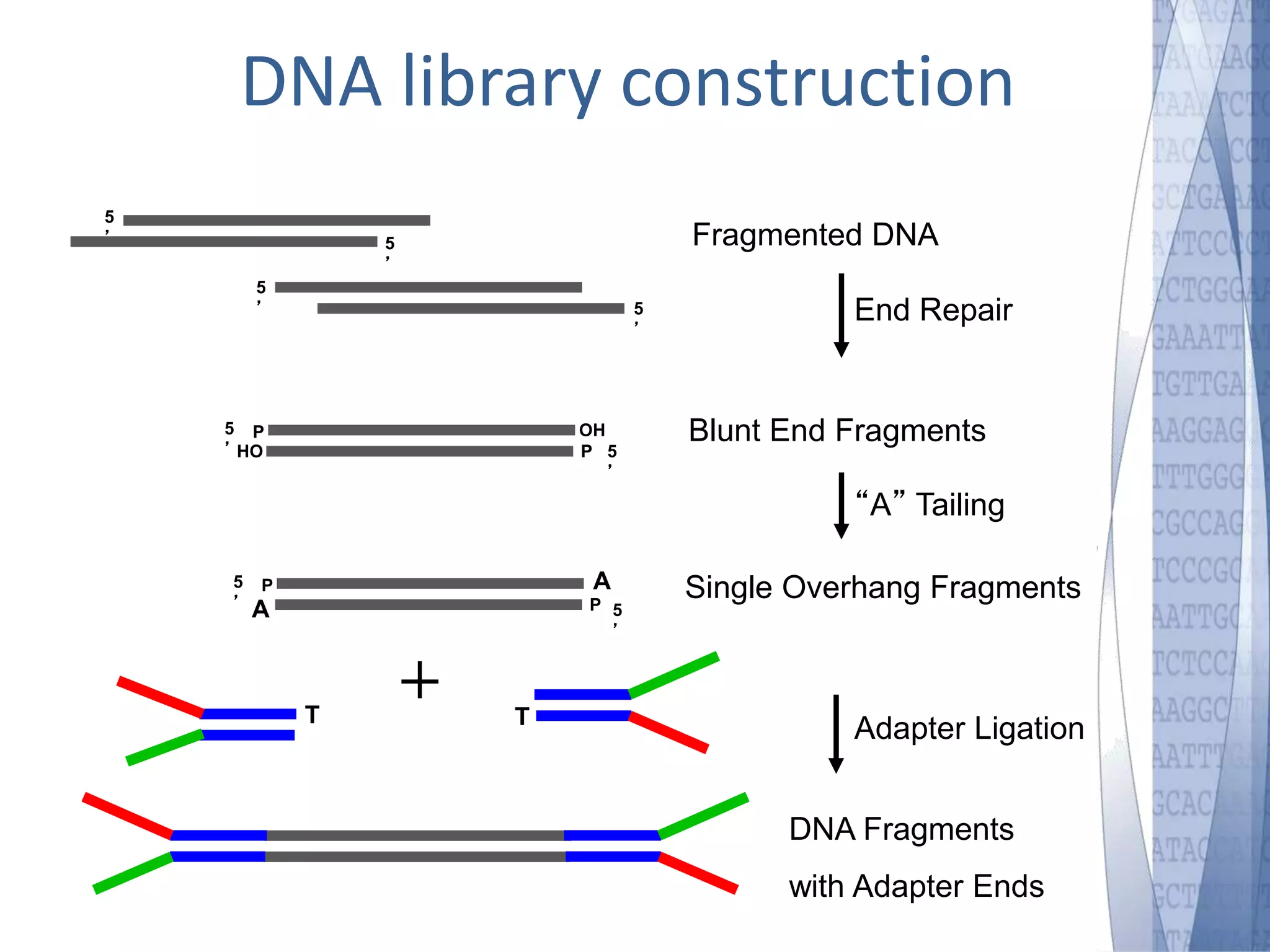
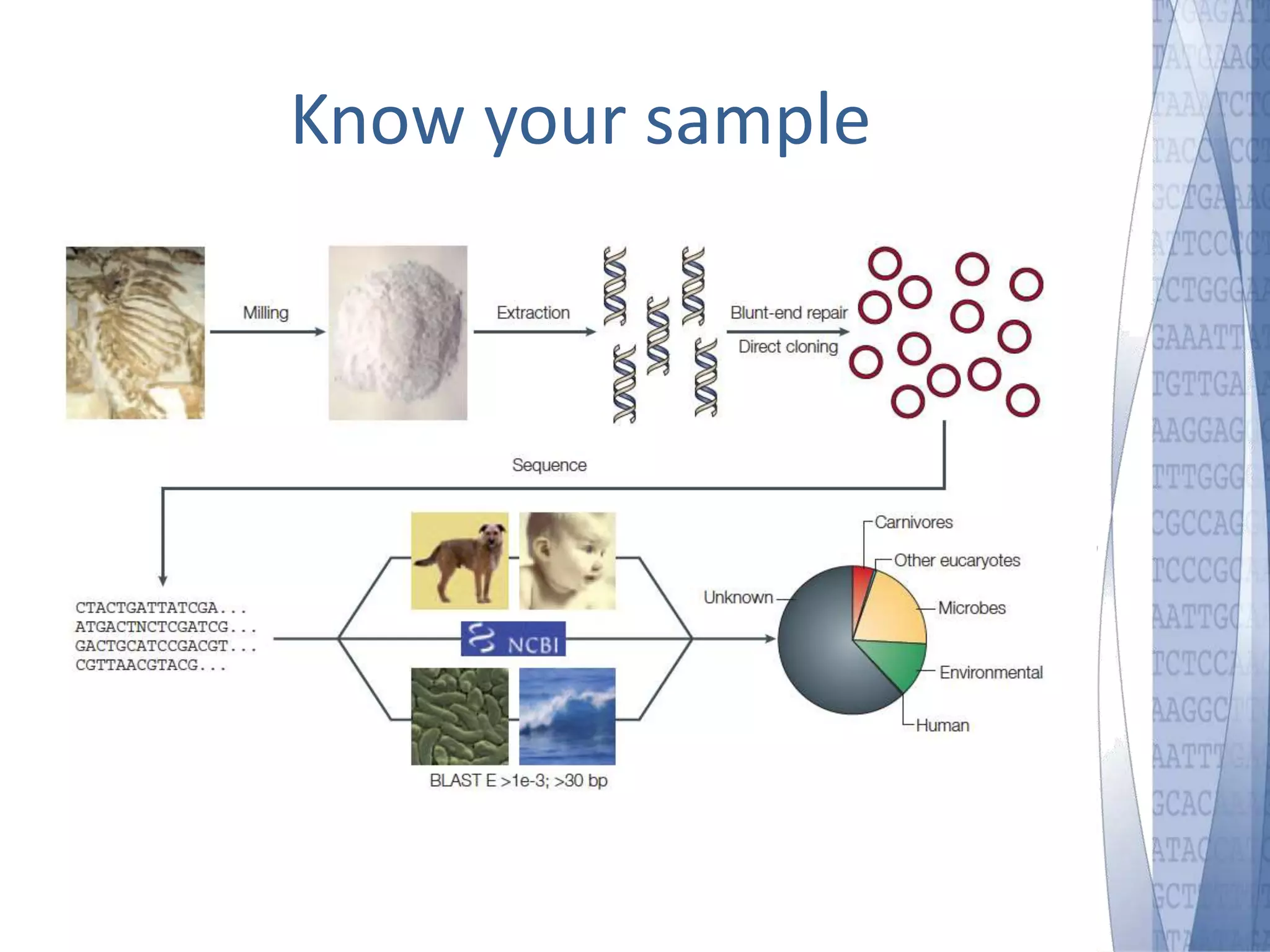
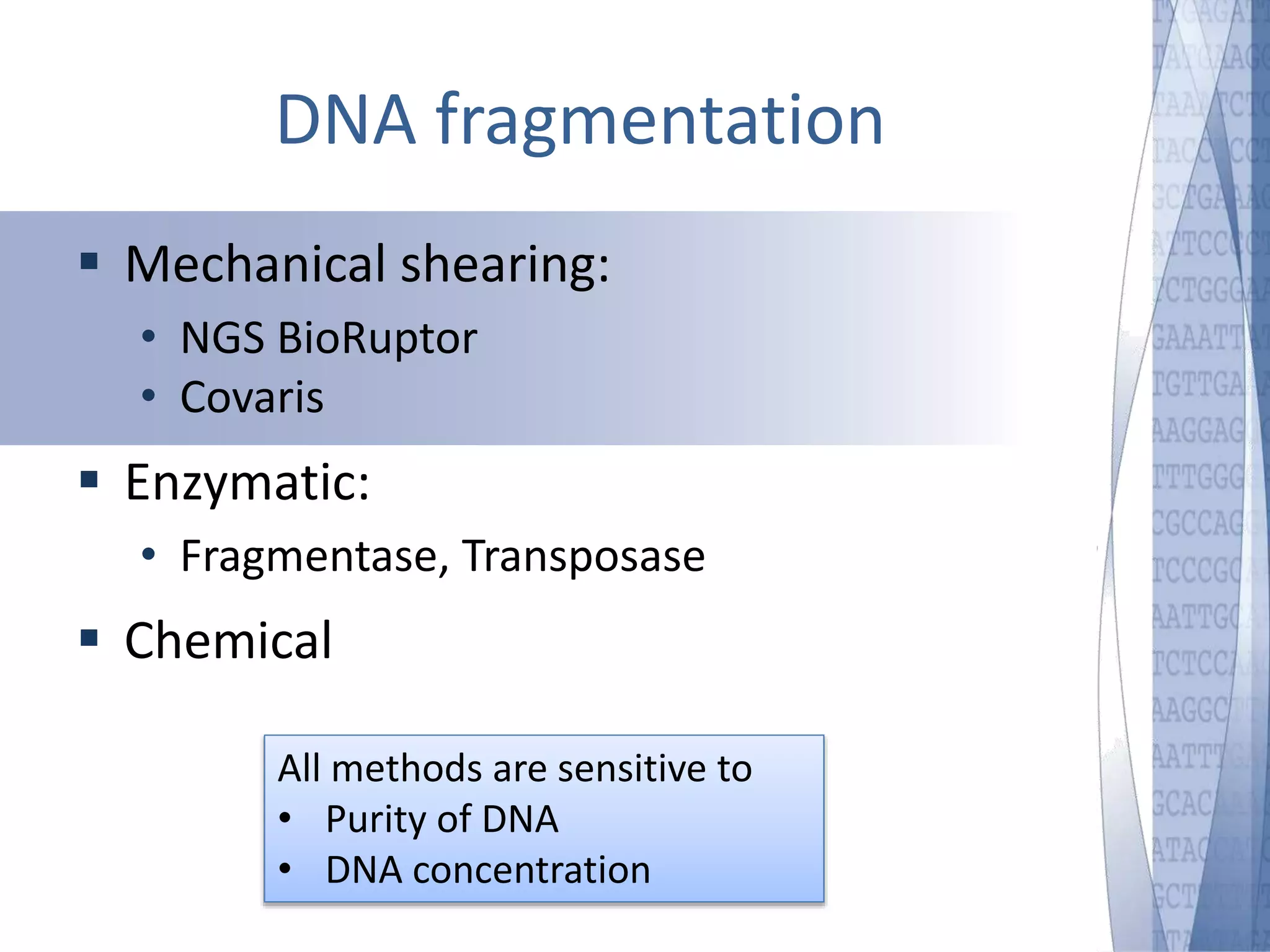
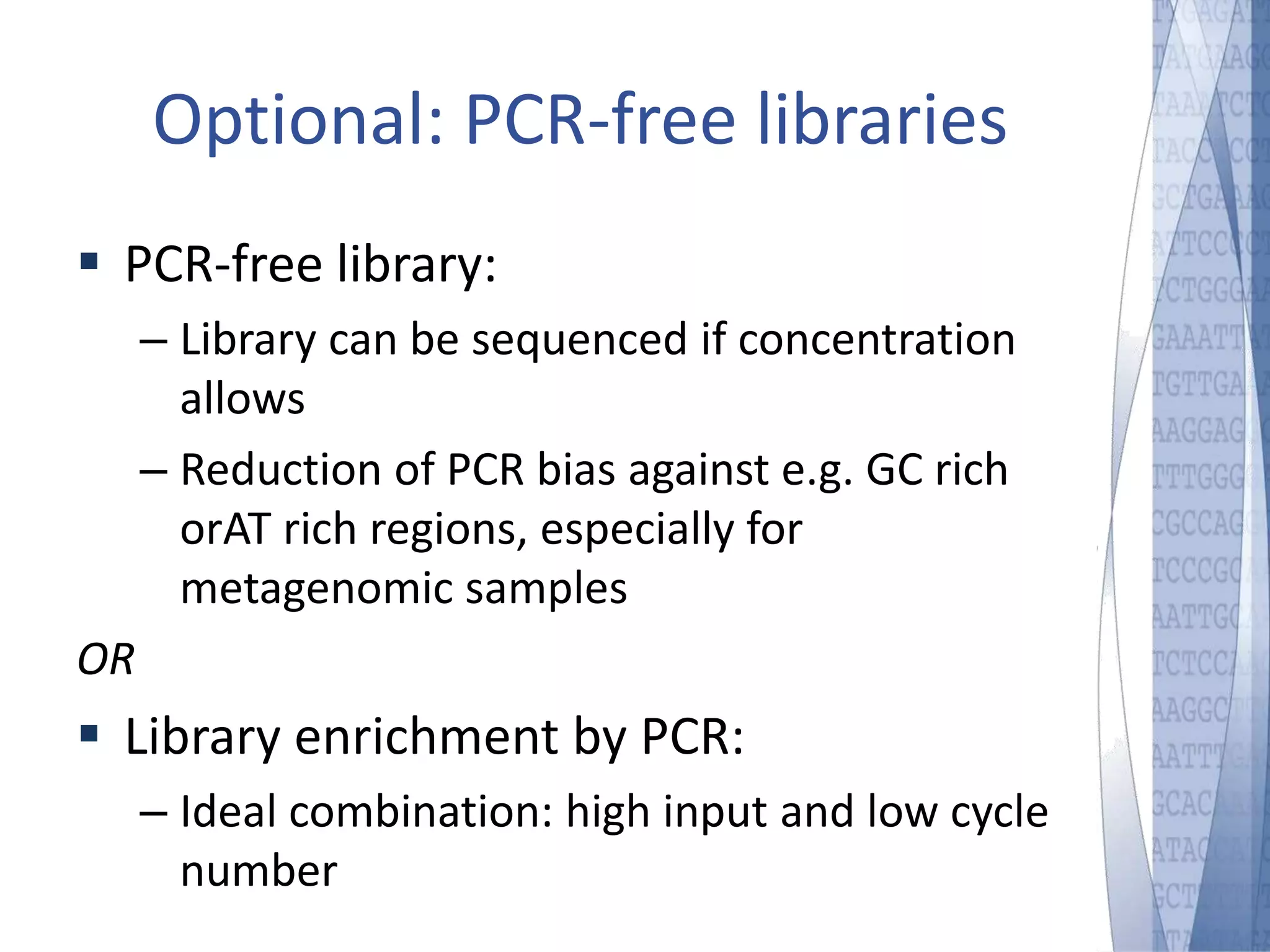
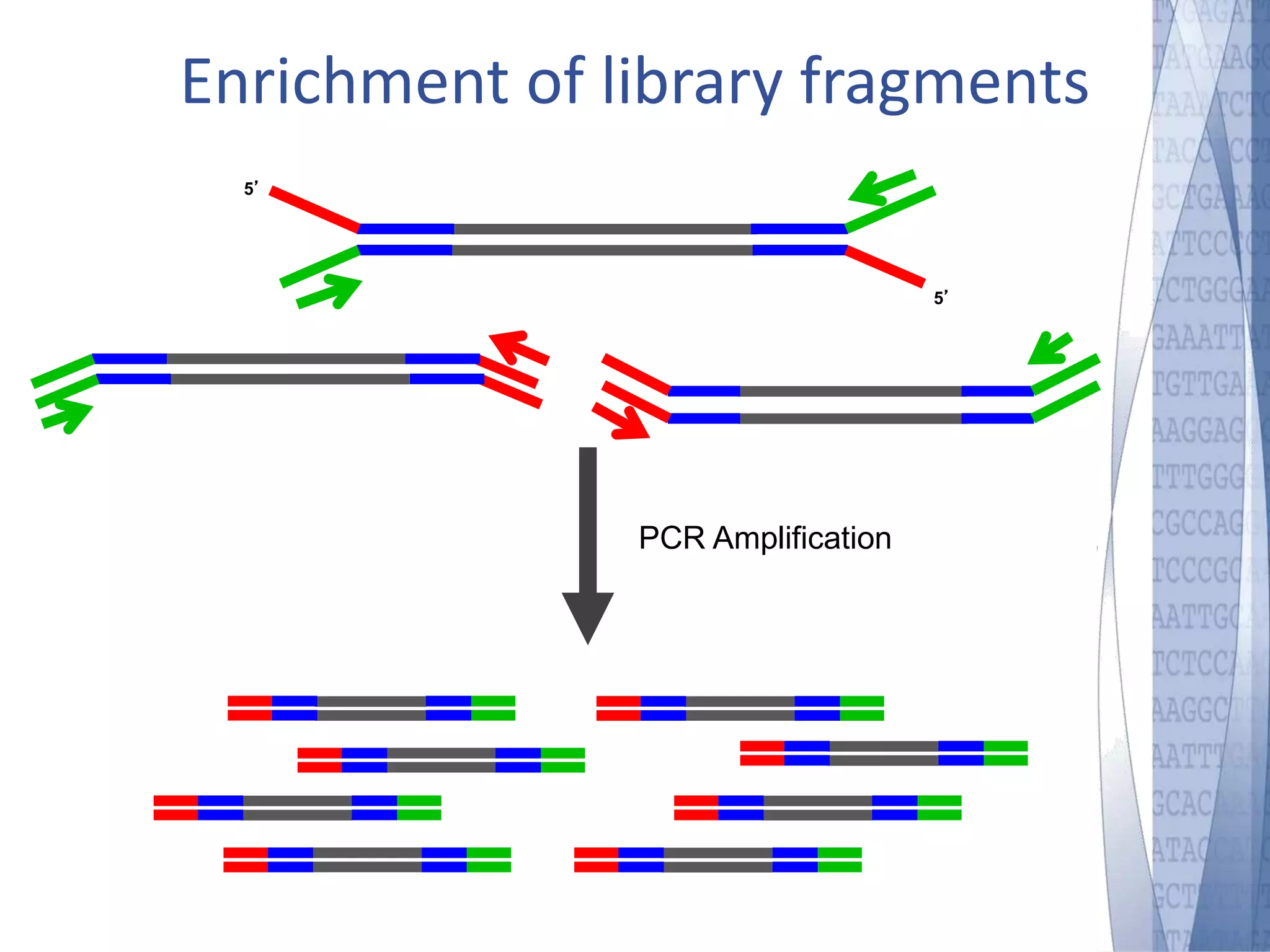
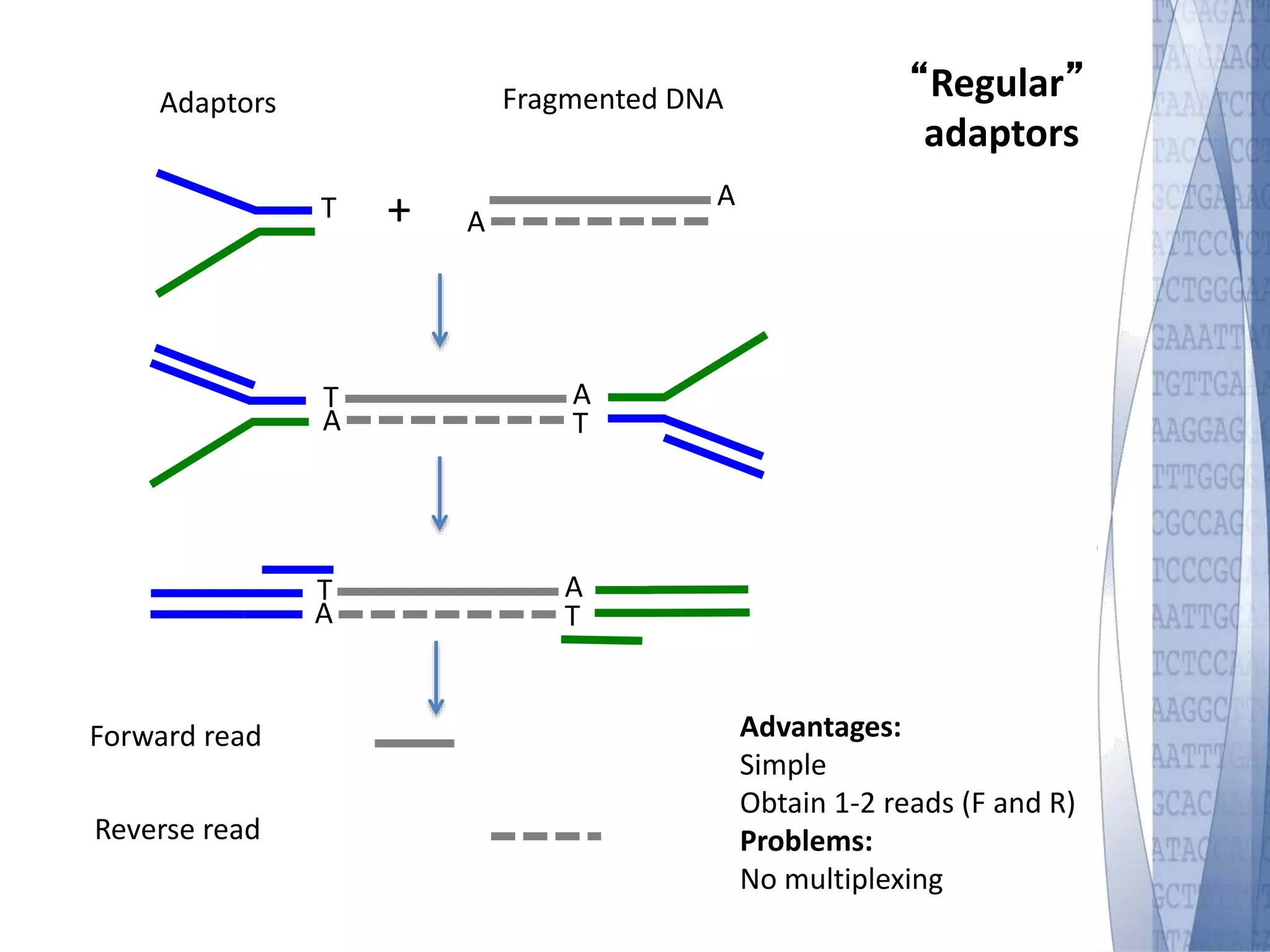
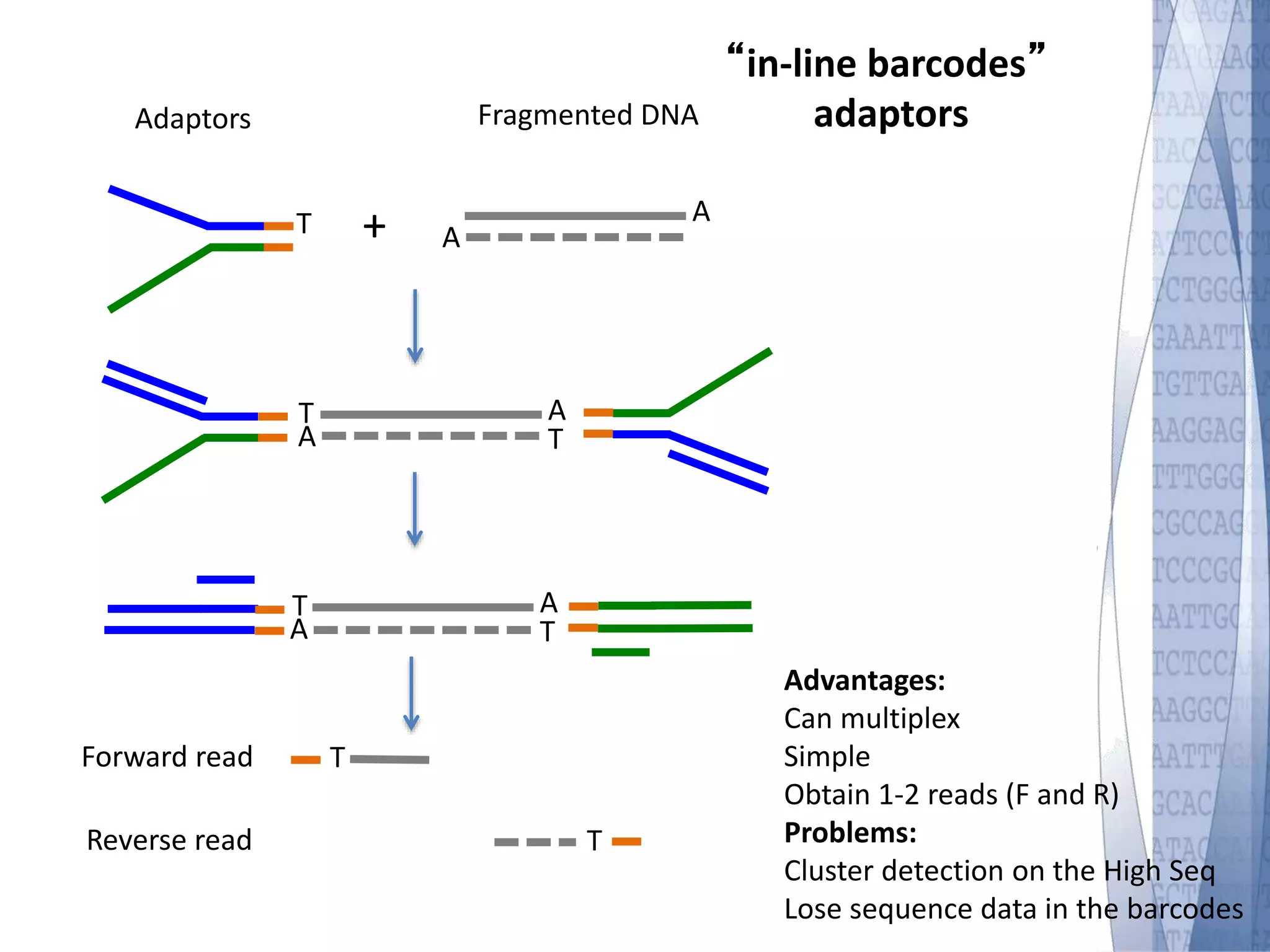
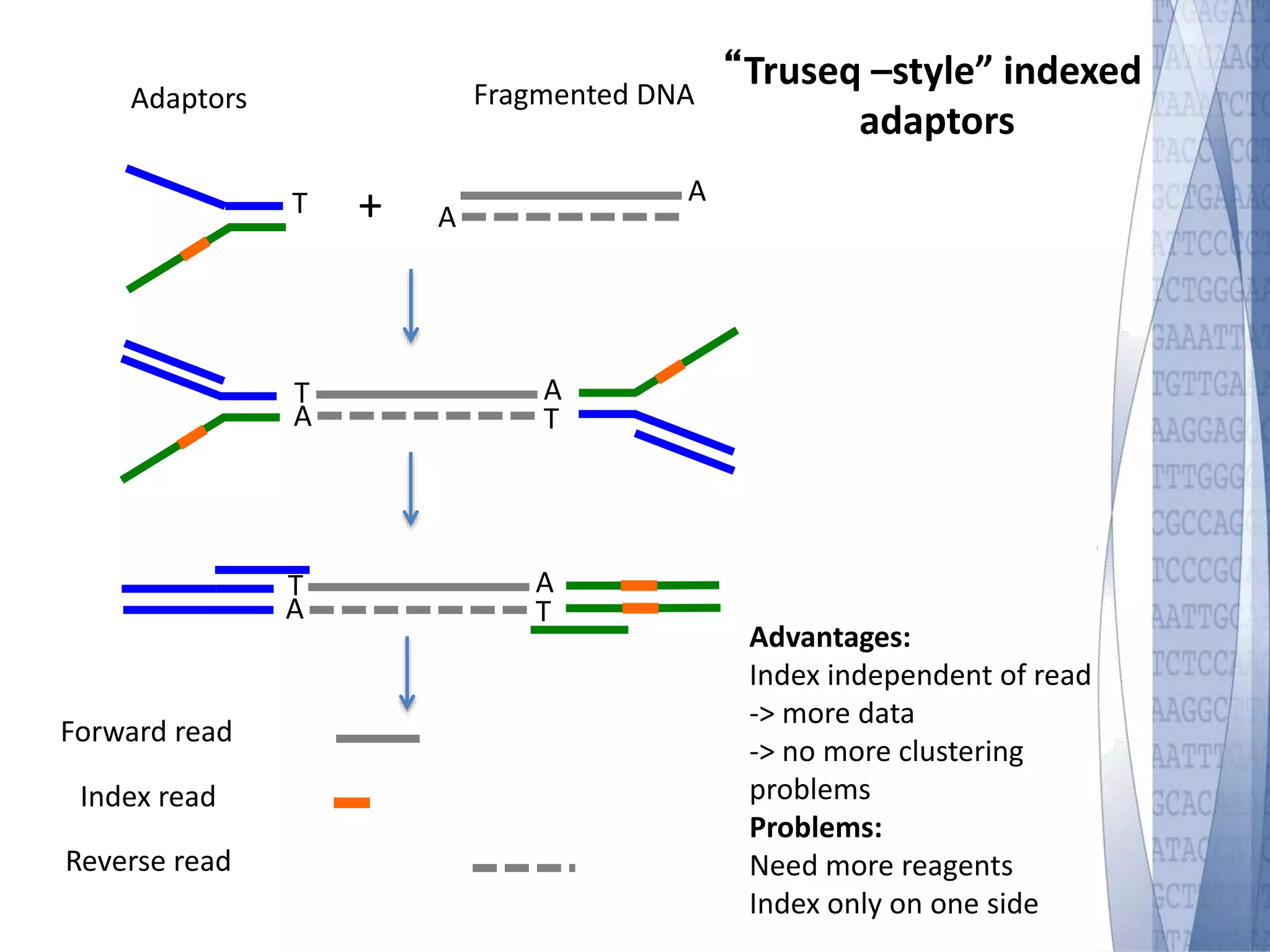
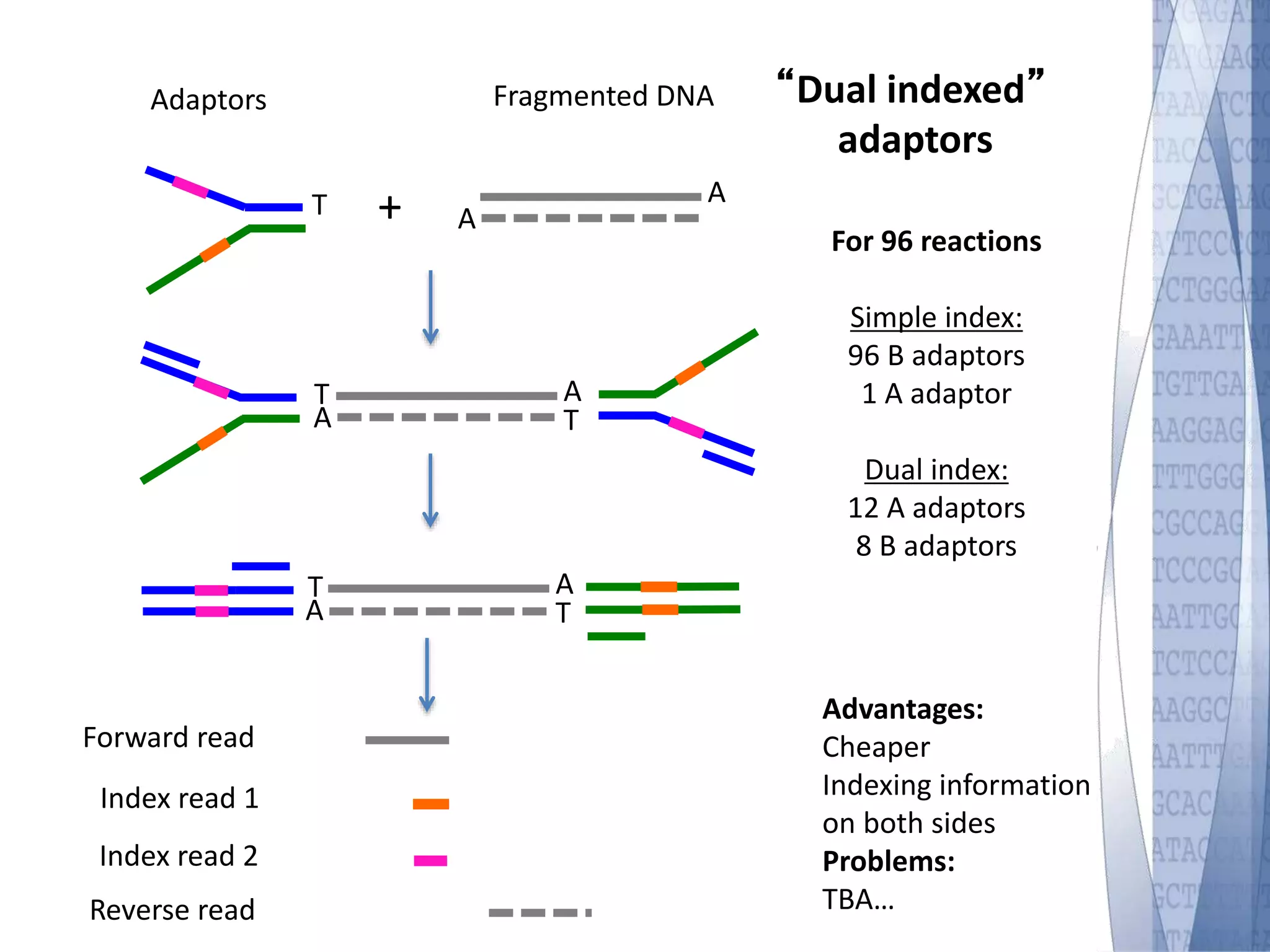
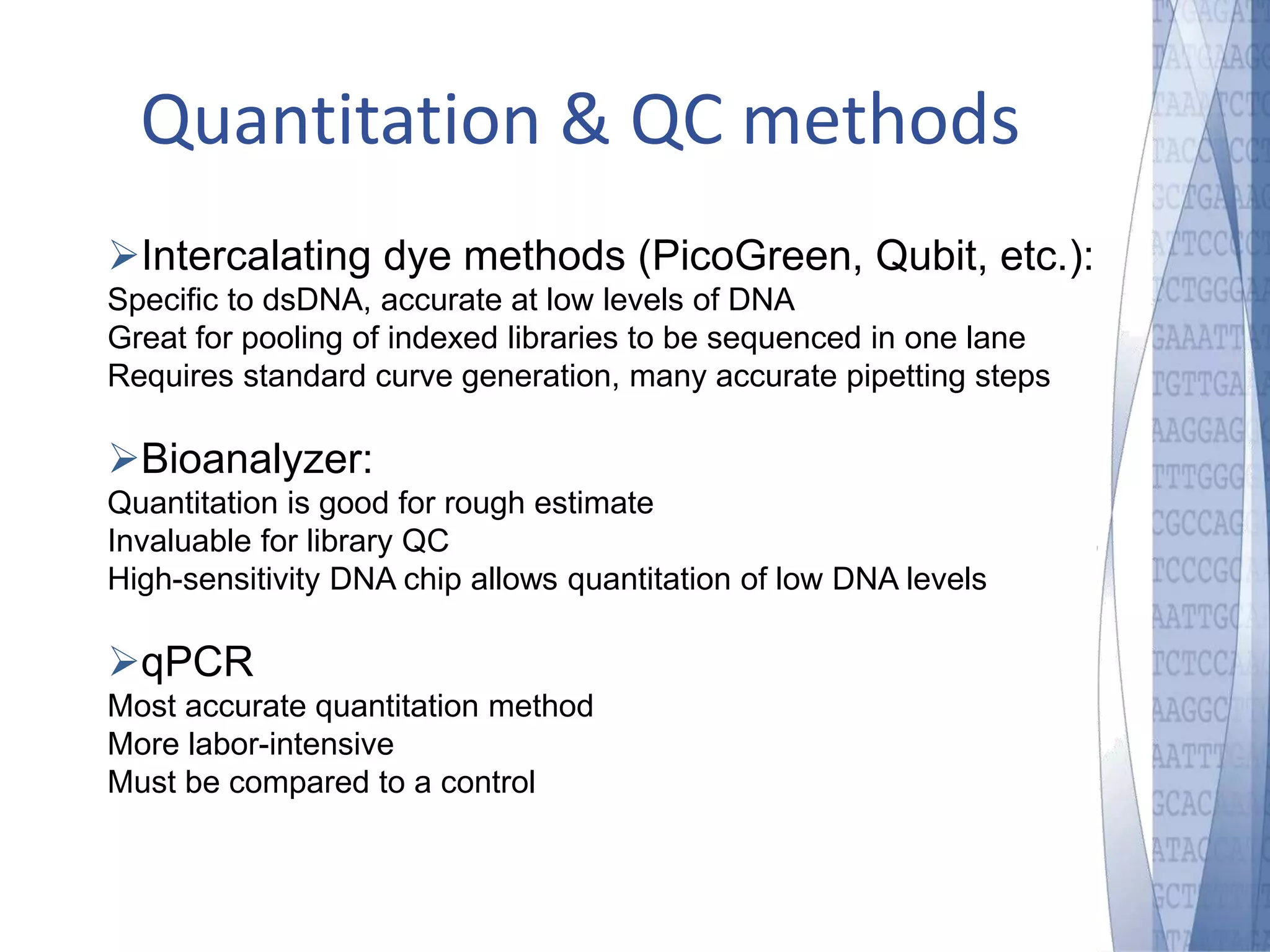
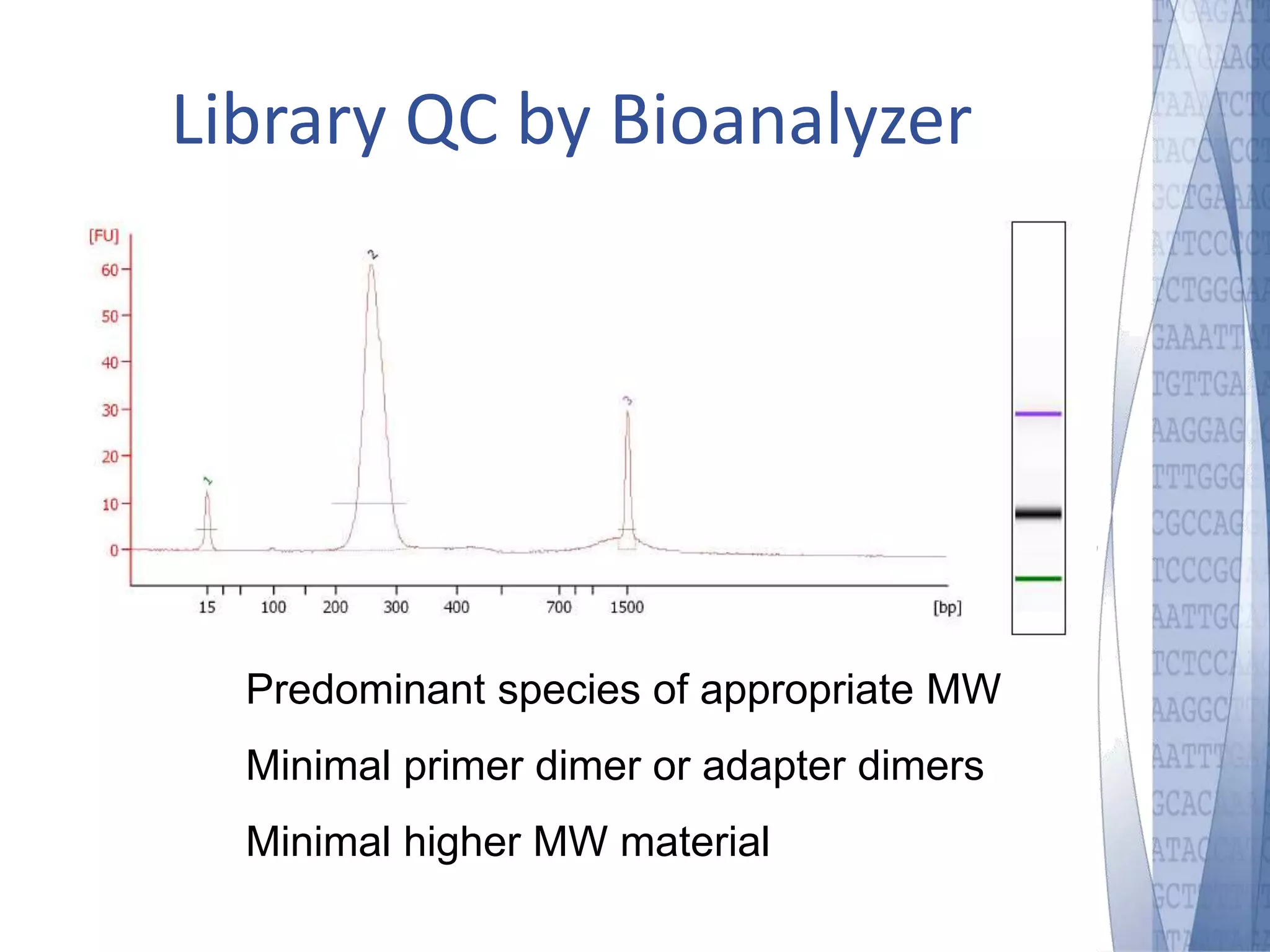
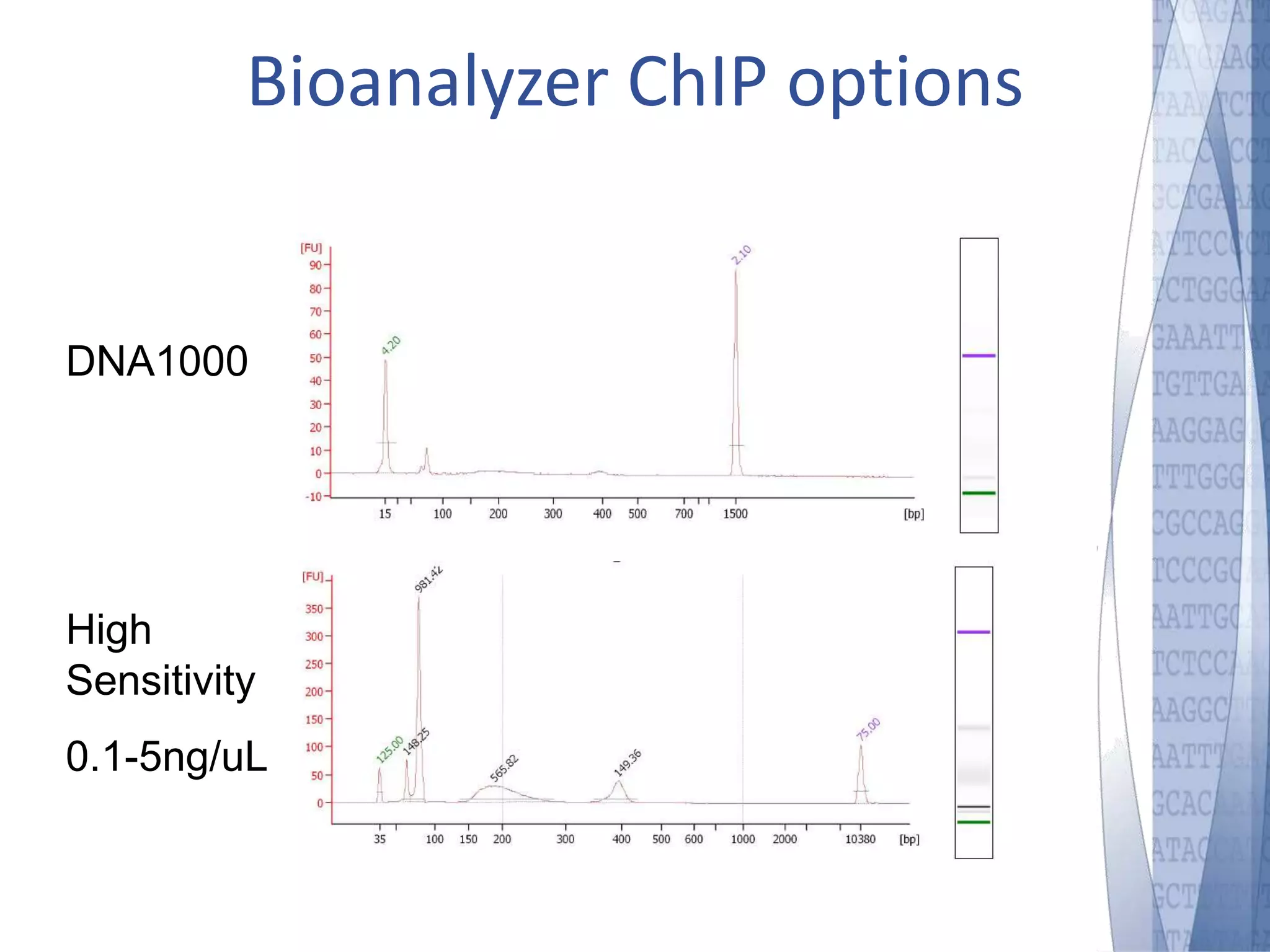
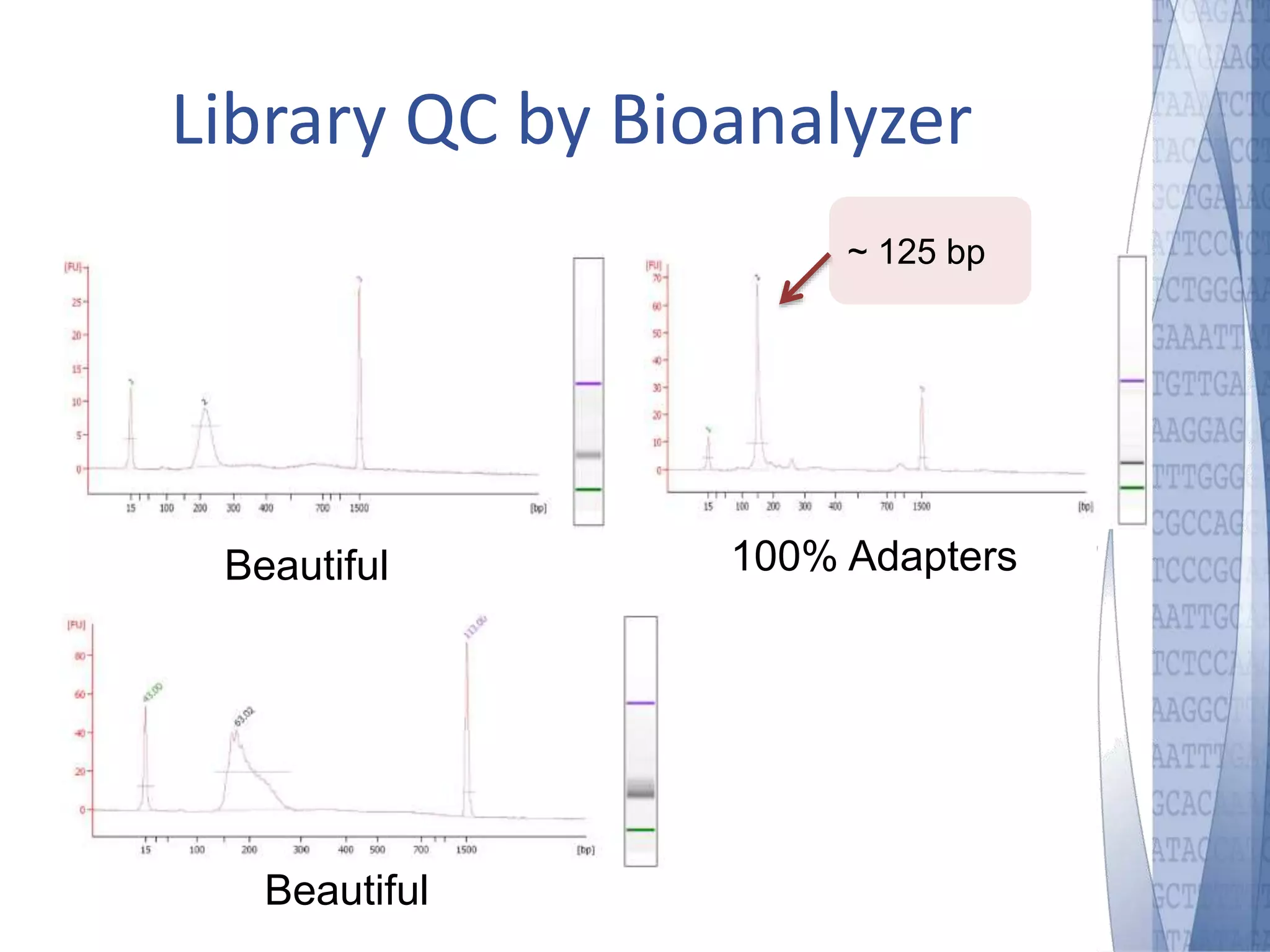
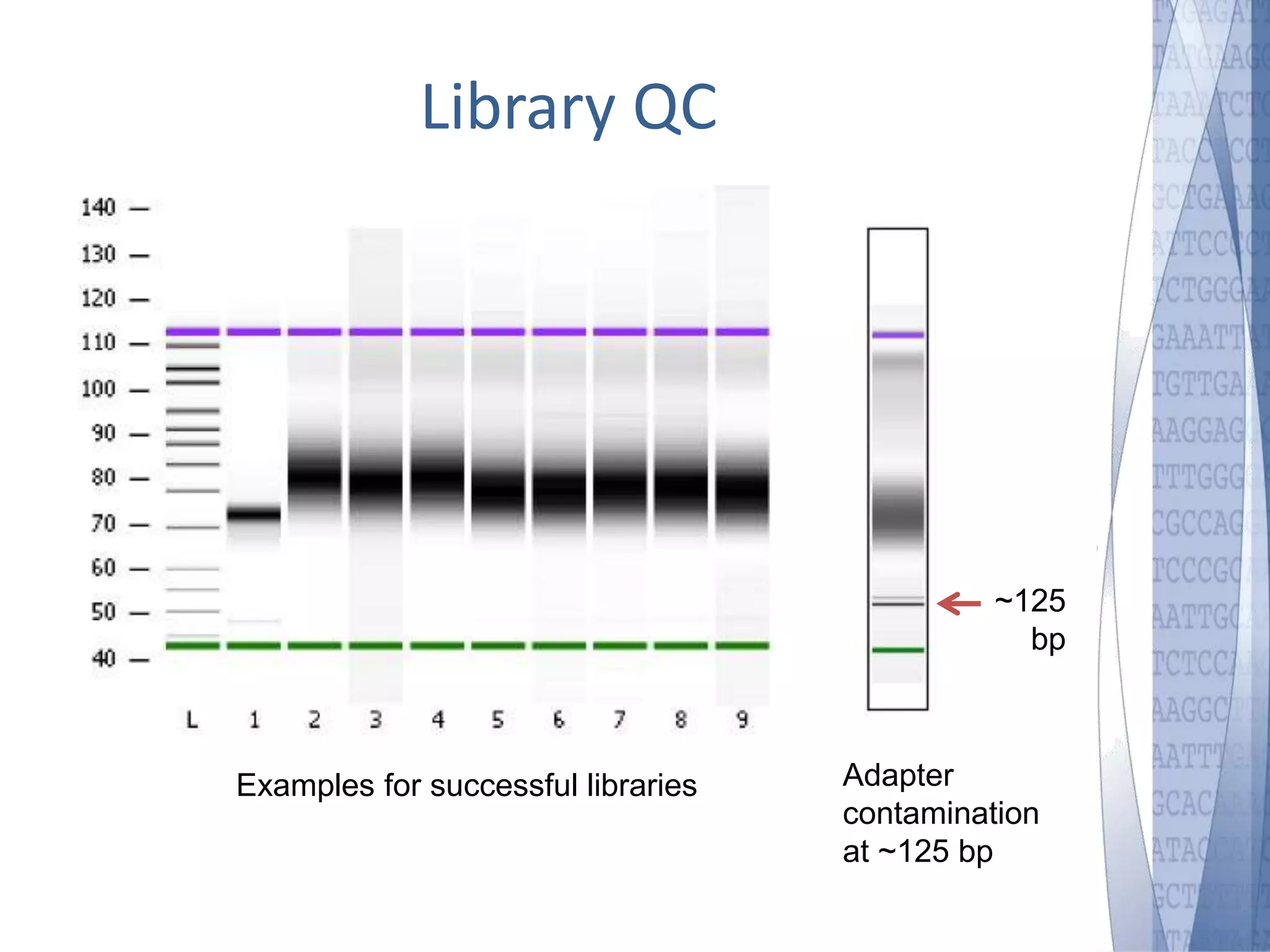
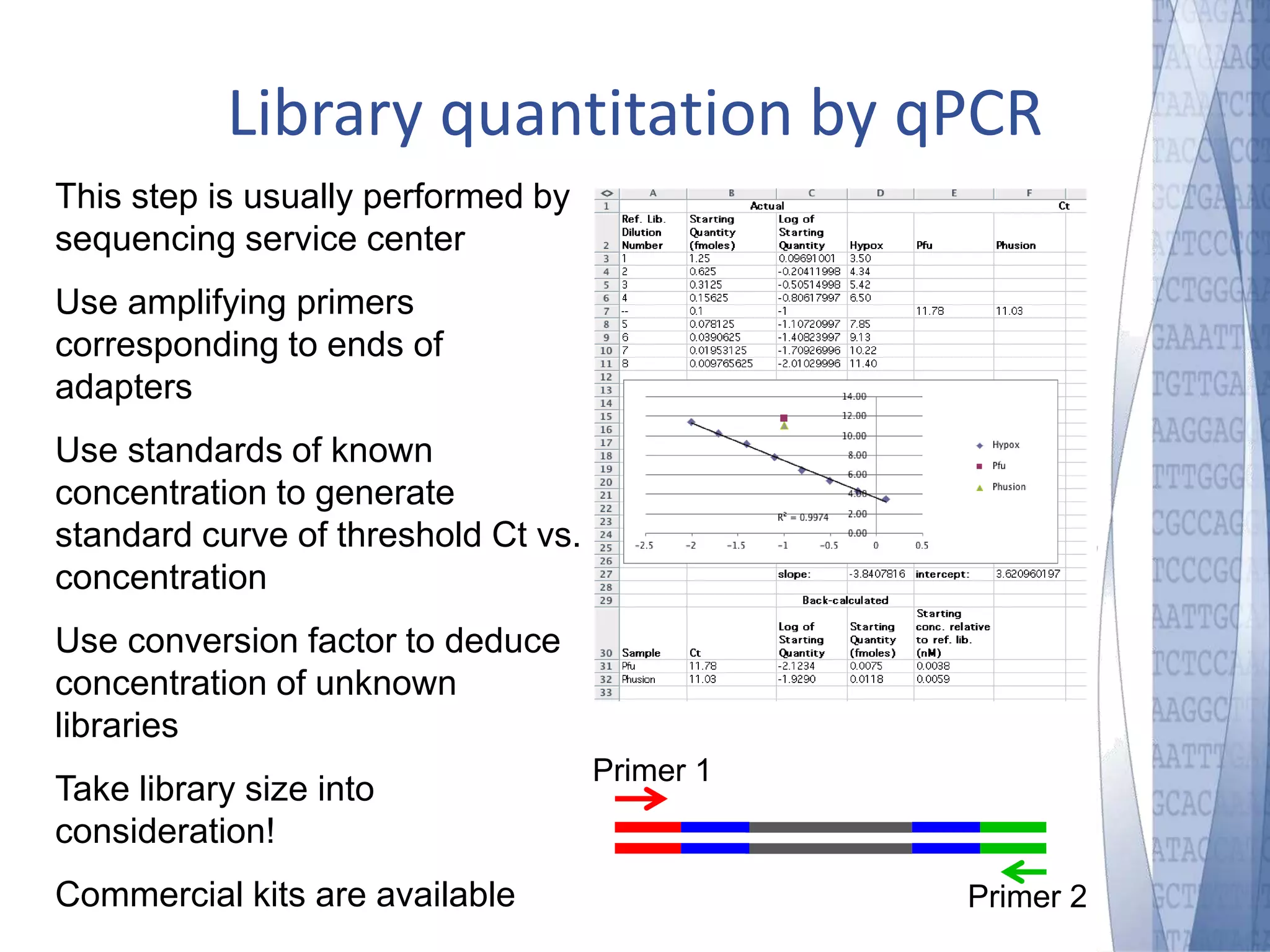
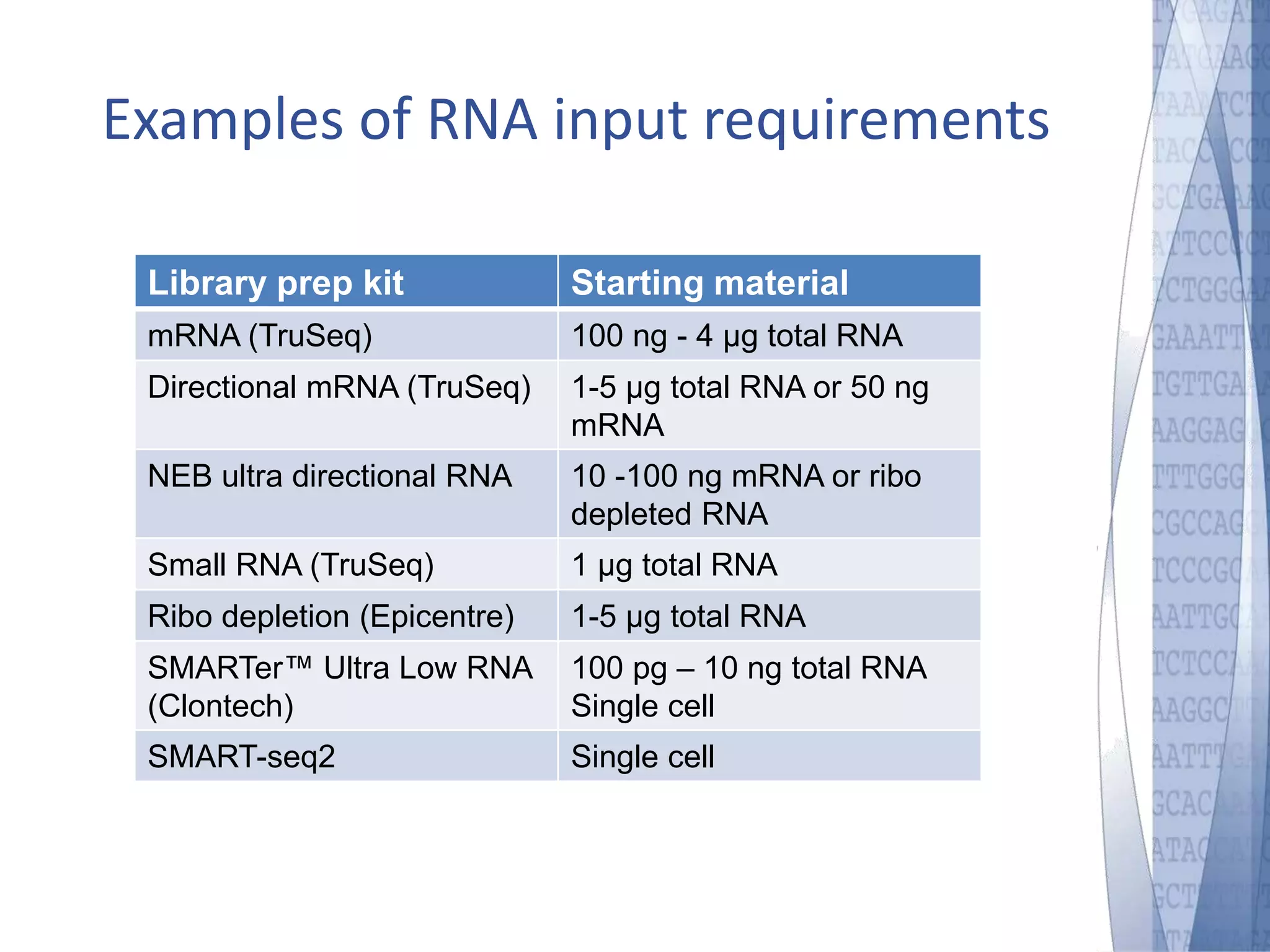
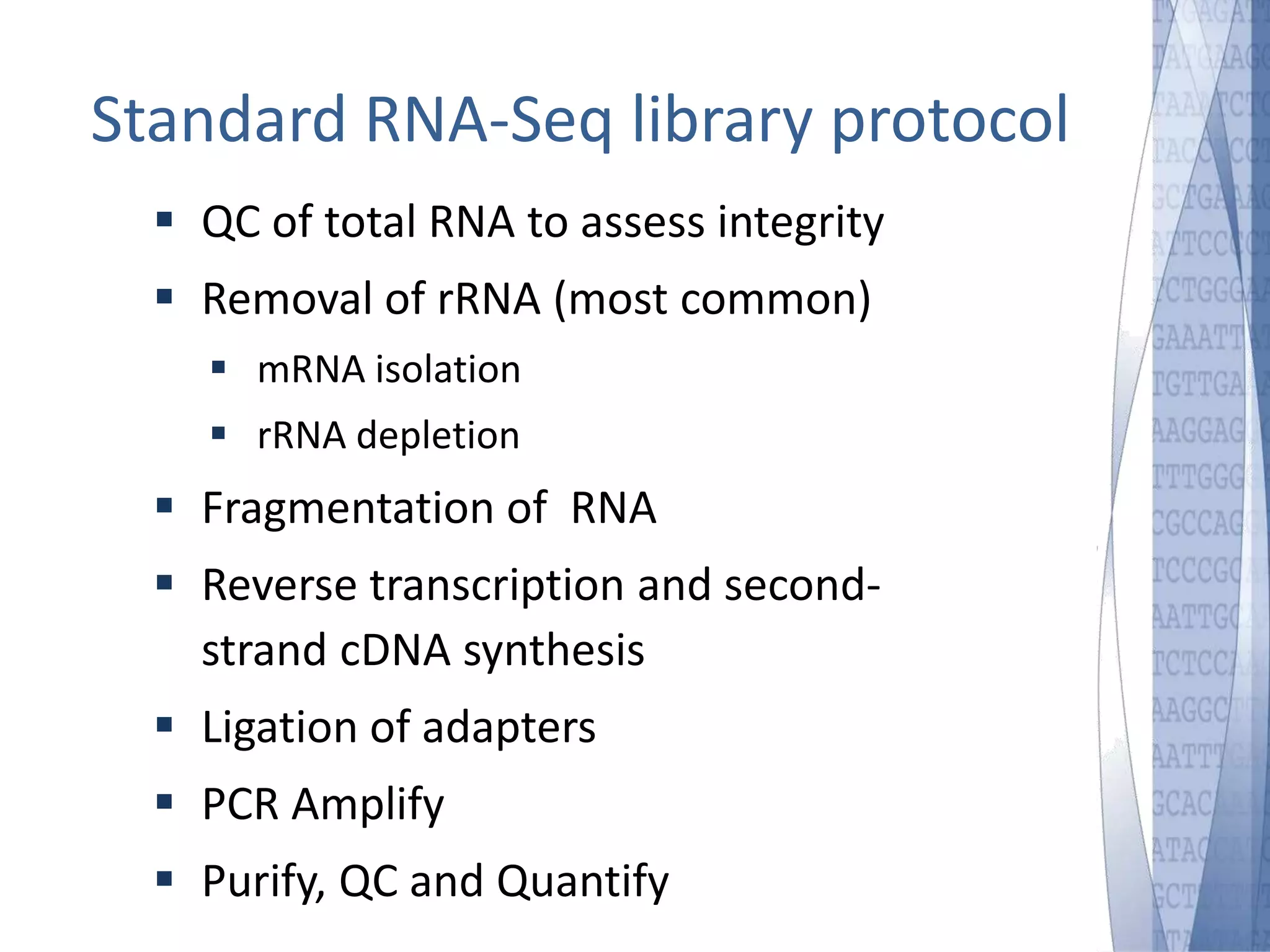
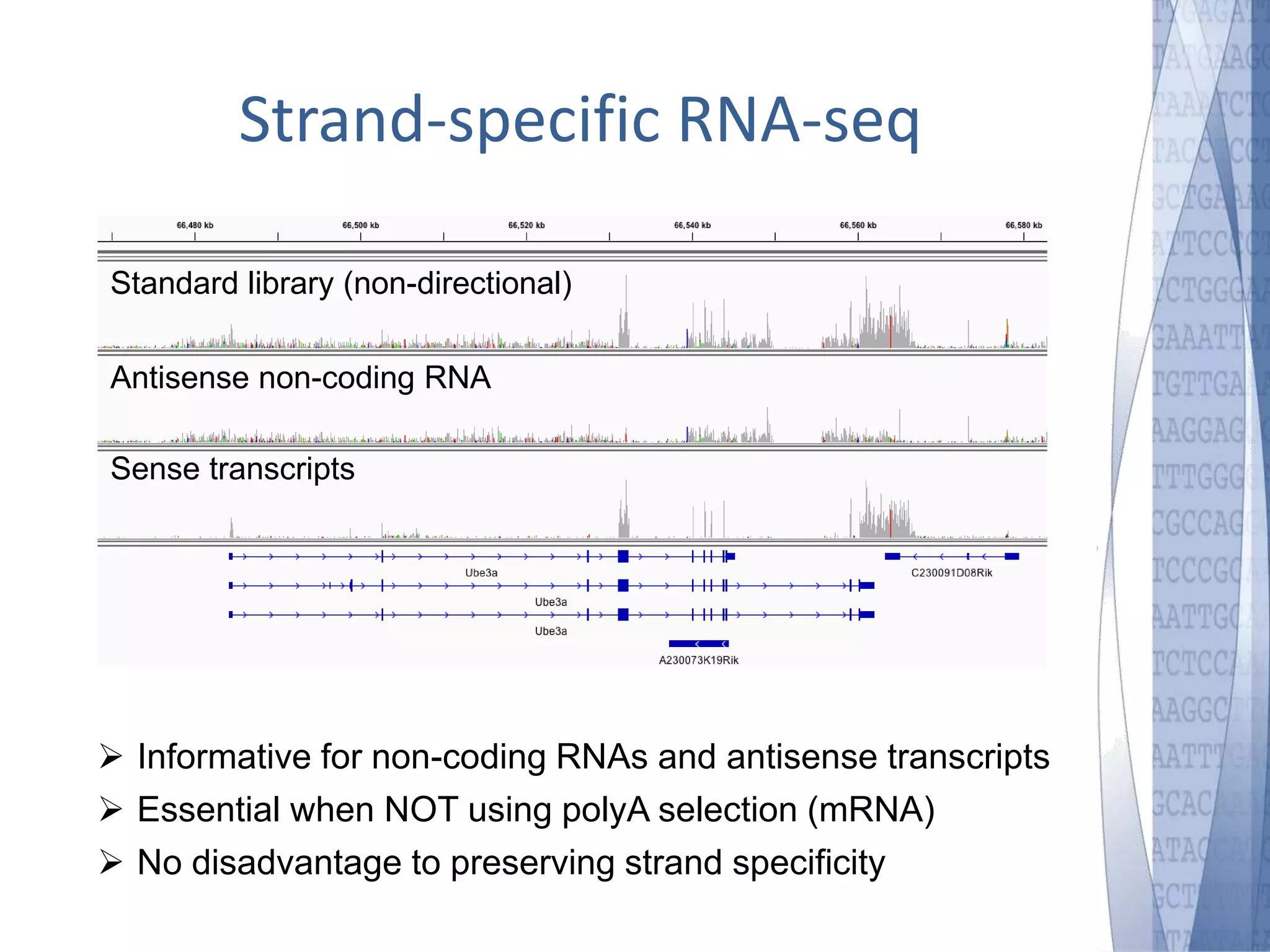
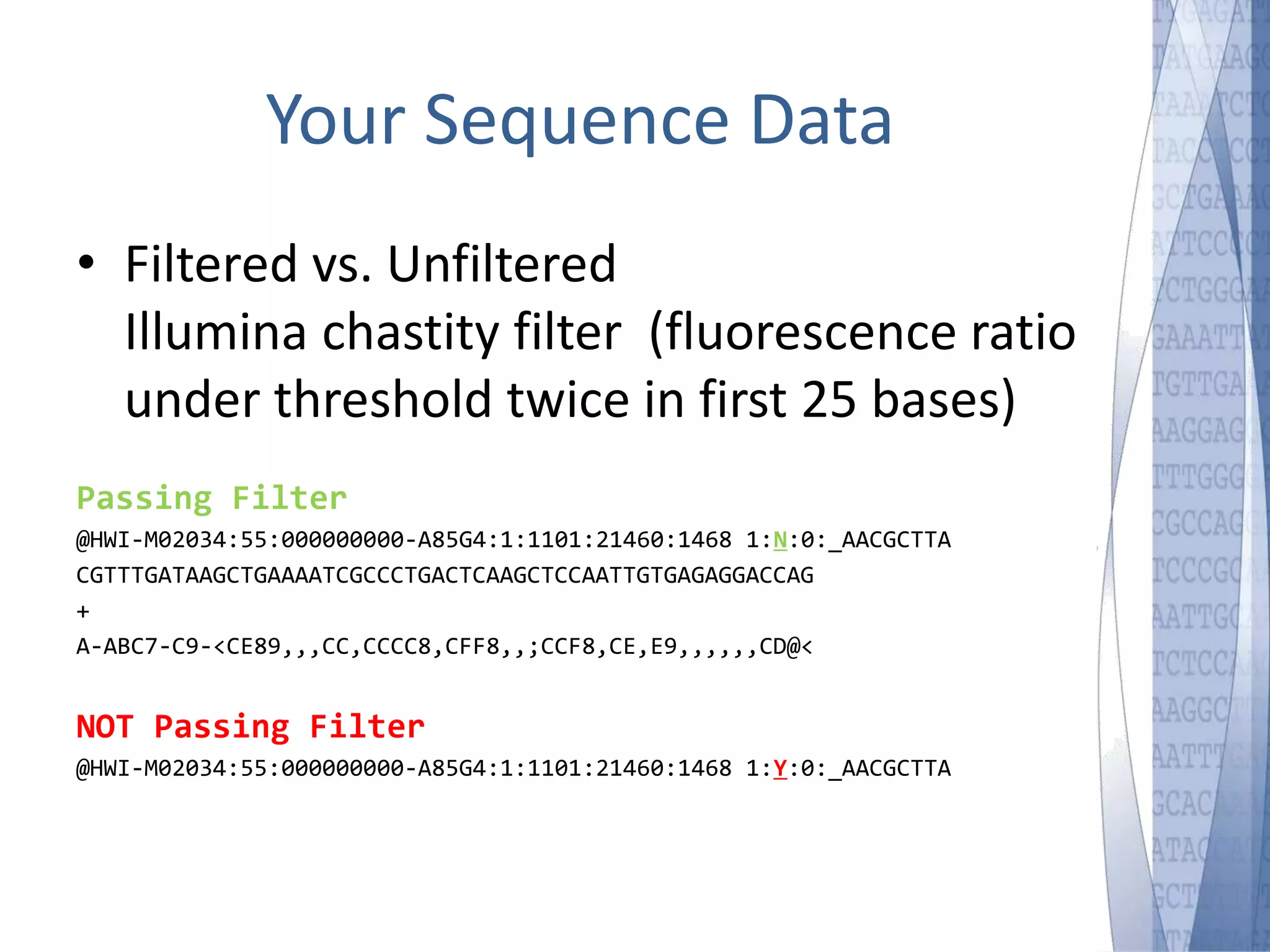
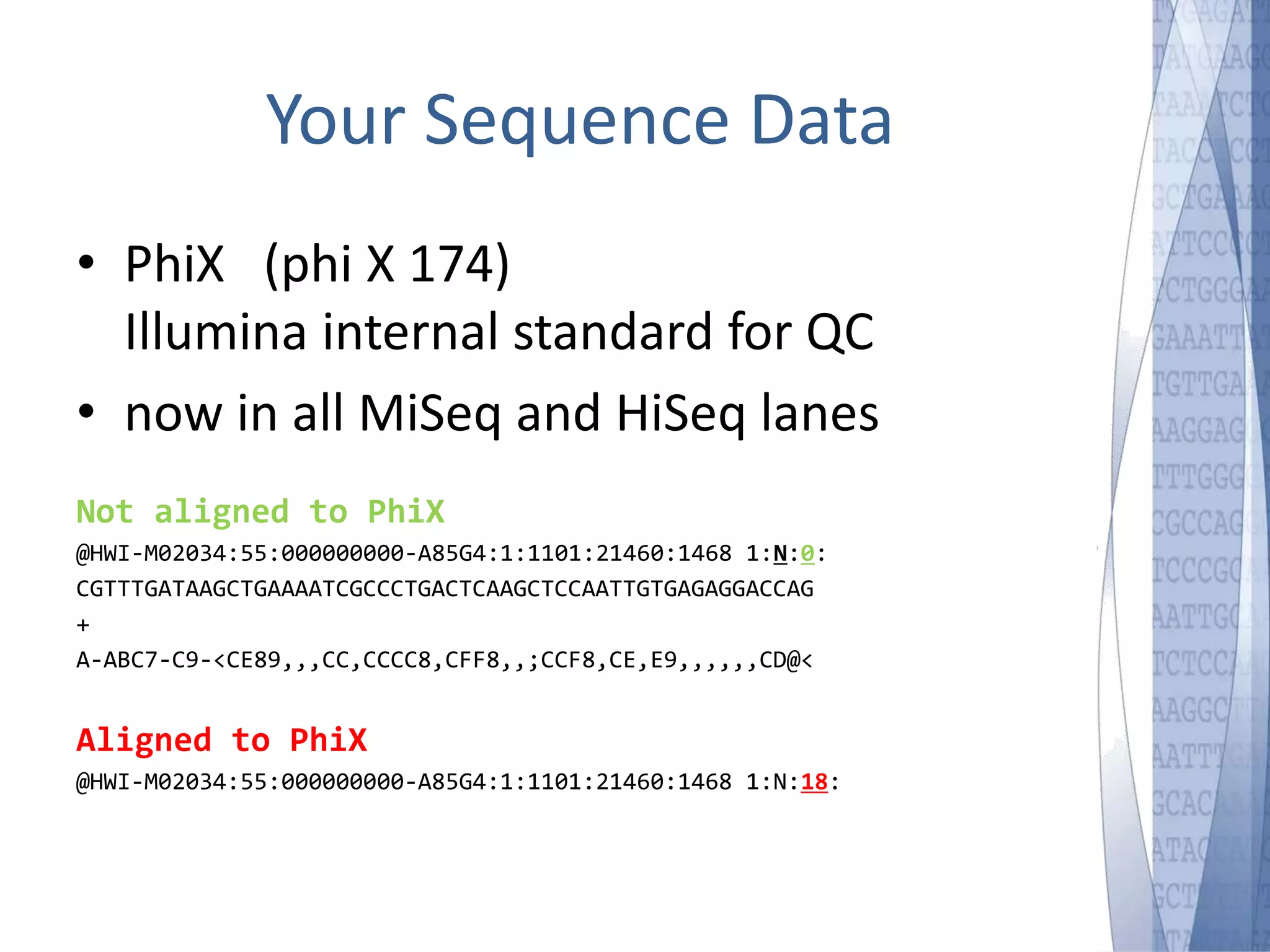
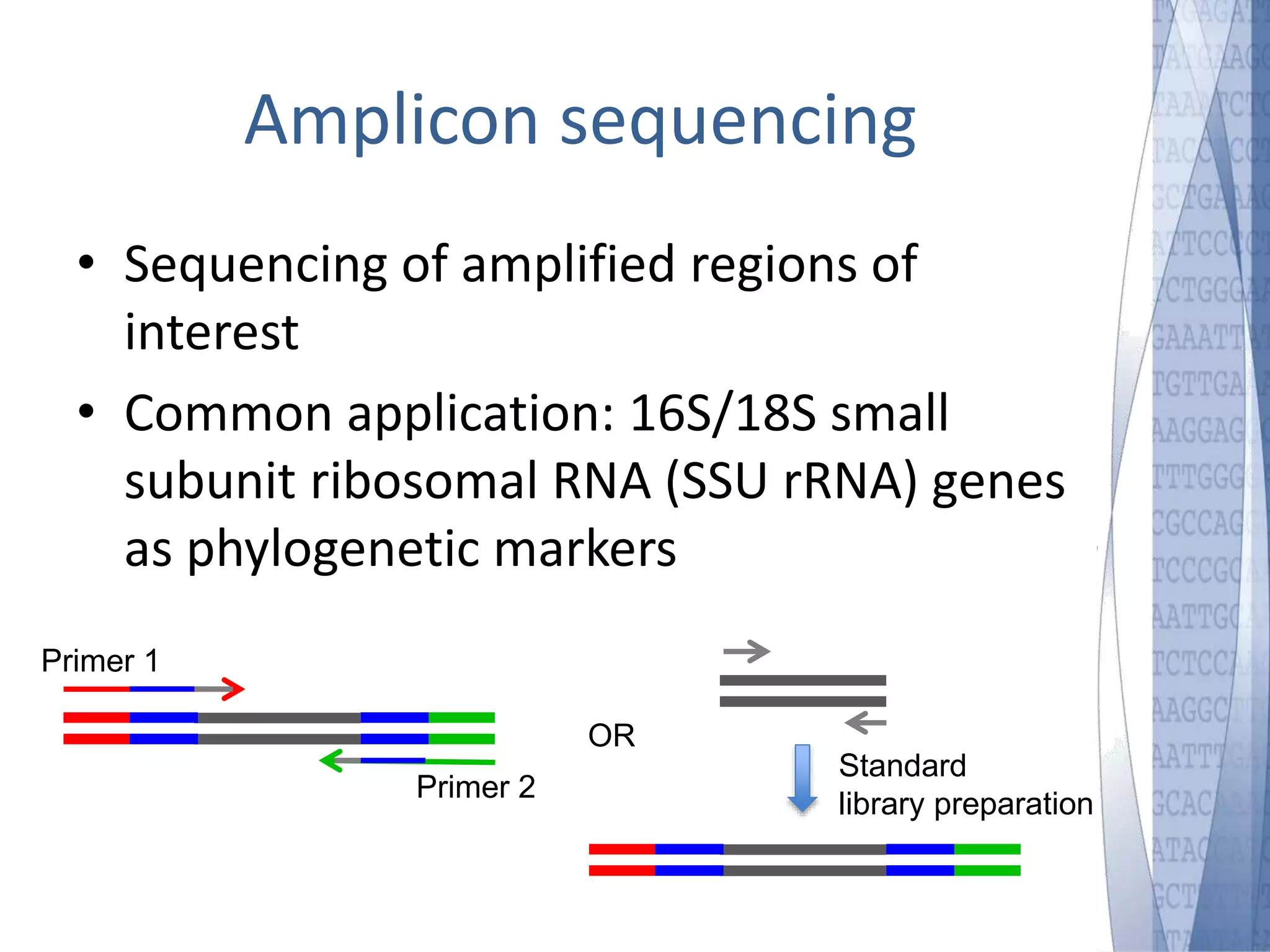
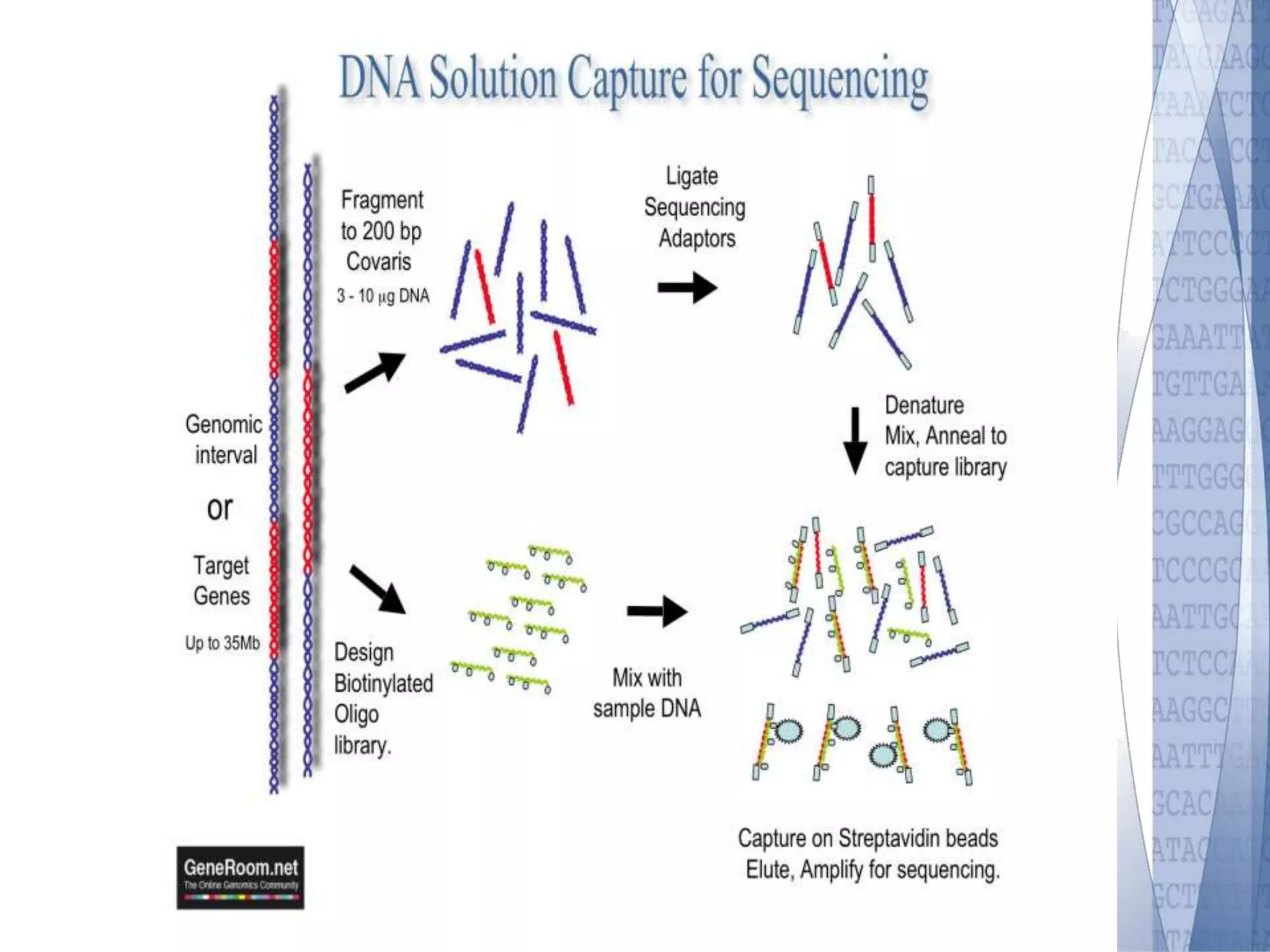
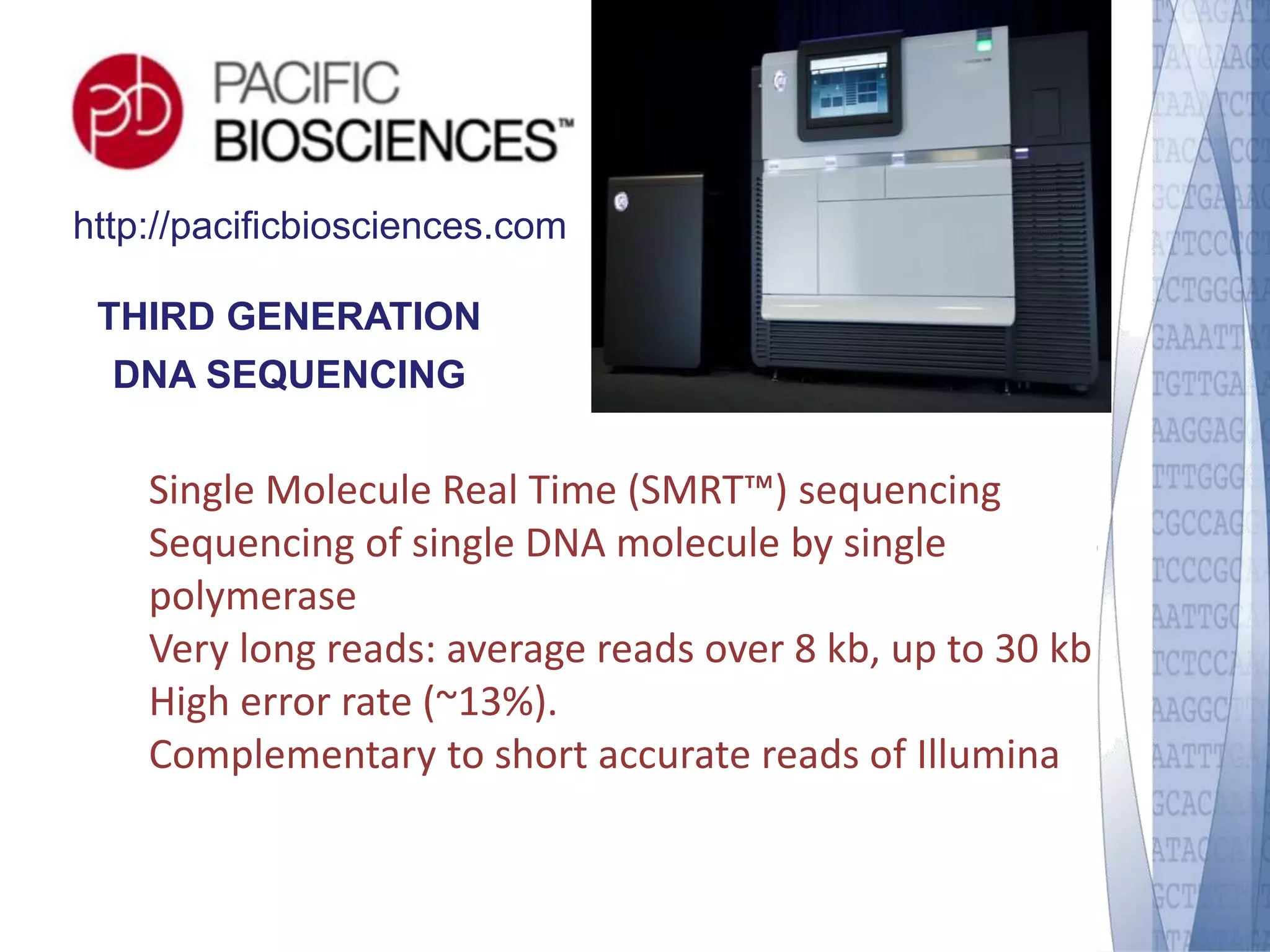
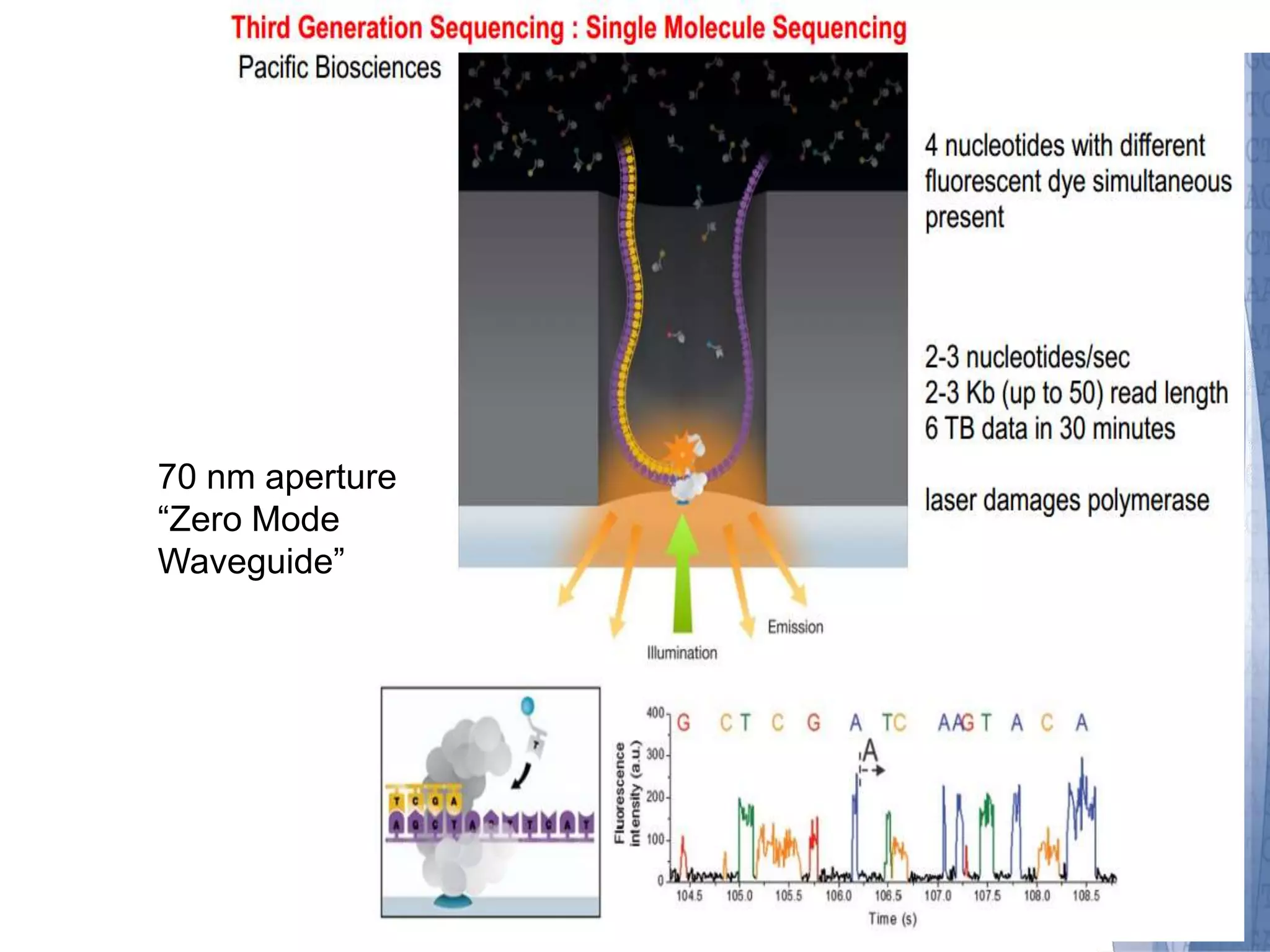
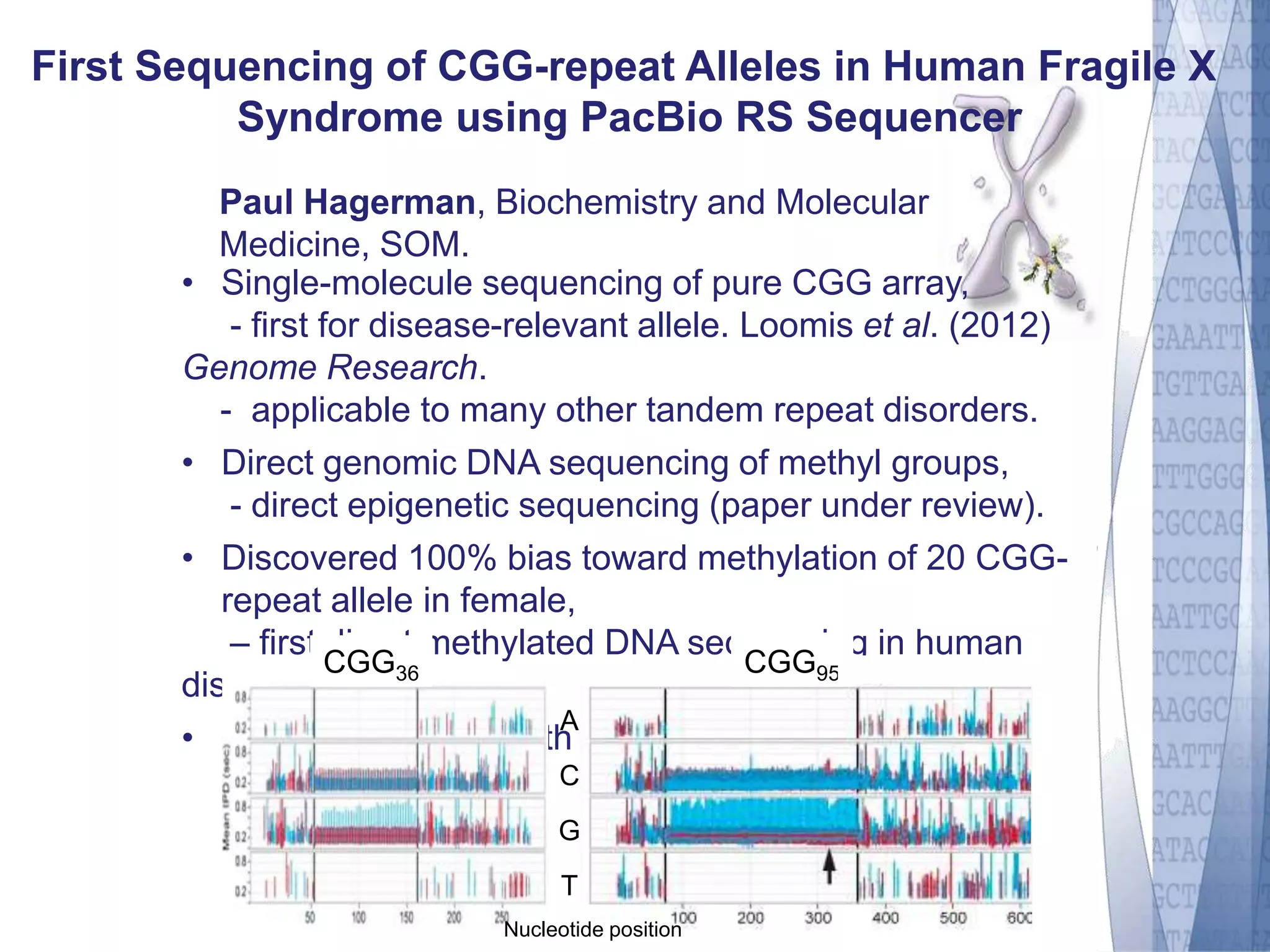
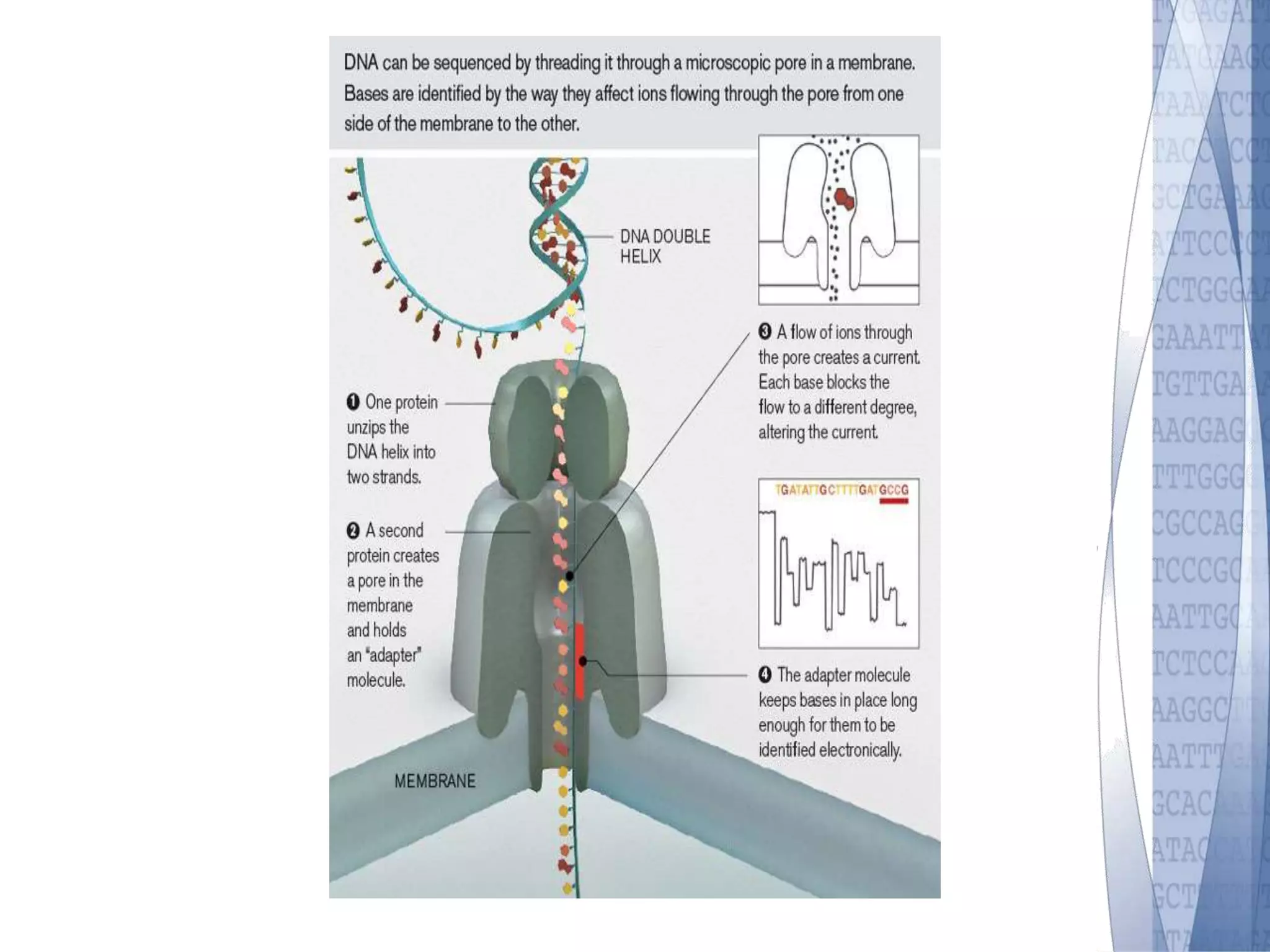
This document discusses wet-lab considerations for Illumina sequencing data analysis. It describes the typical Illumina sequencing workflow including library preparation, cluster formation, sequencing, and data analysis. It provides details on DNA and RNA input requirements, library construction steps like fragmentation and adapter ligation, and quality control methods. The document also discusses newer sequencing technologies like Pacific Biosciences and Oxford Nanopore sequencing.