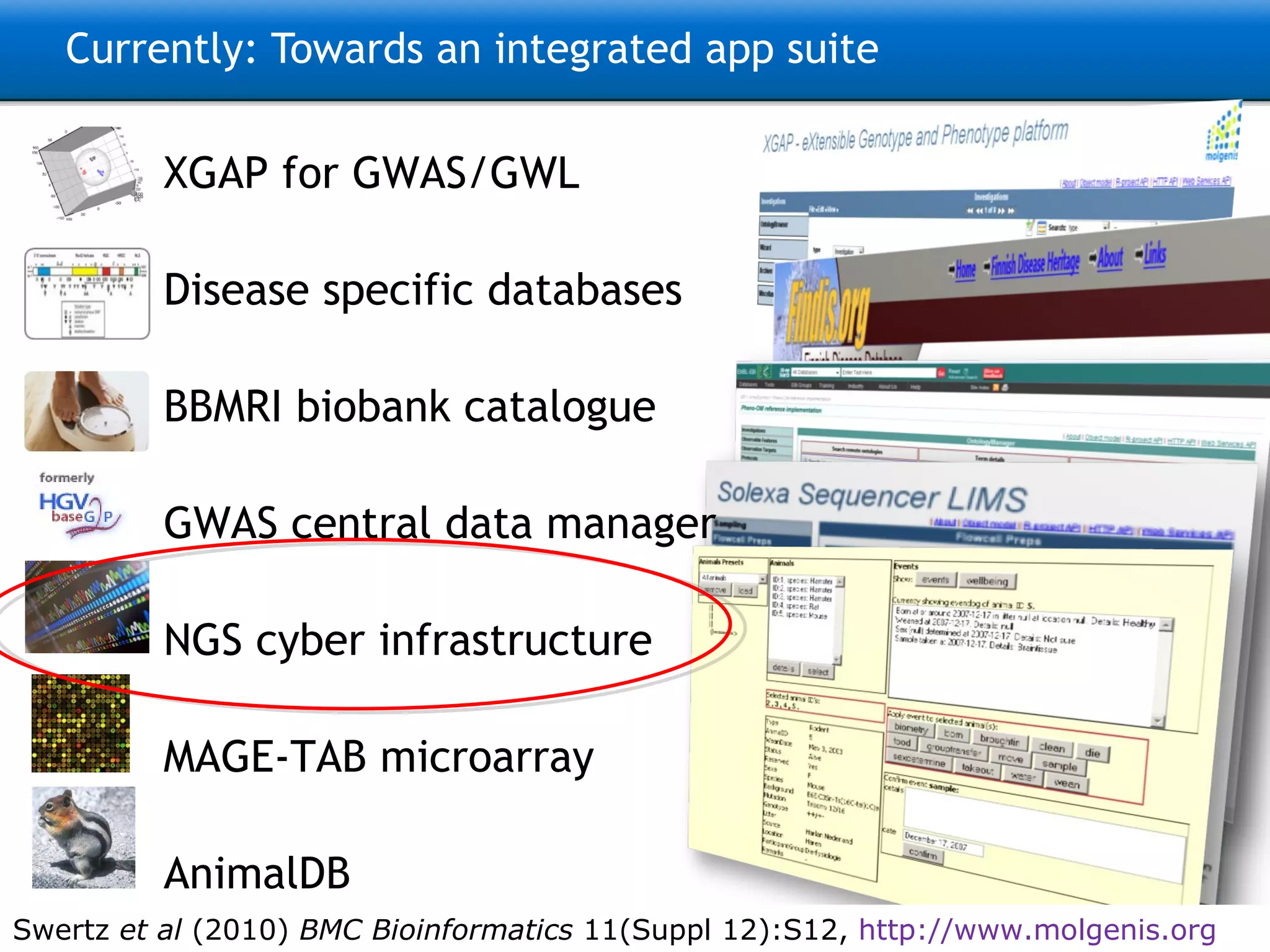
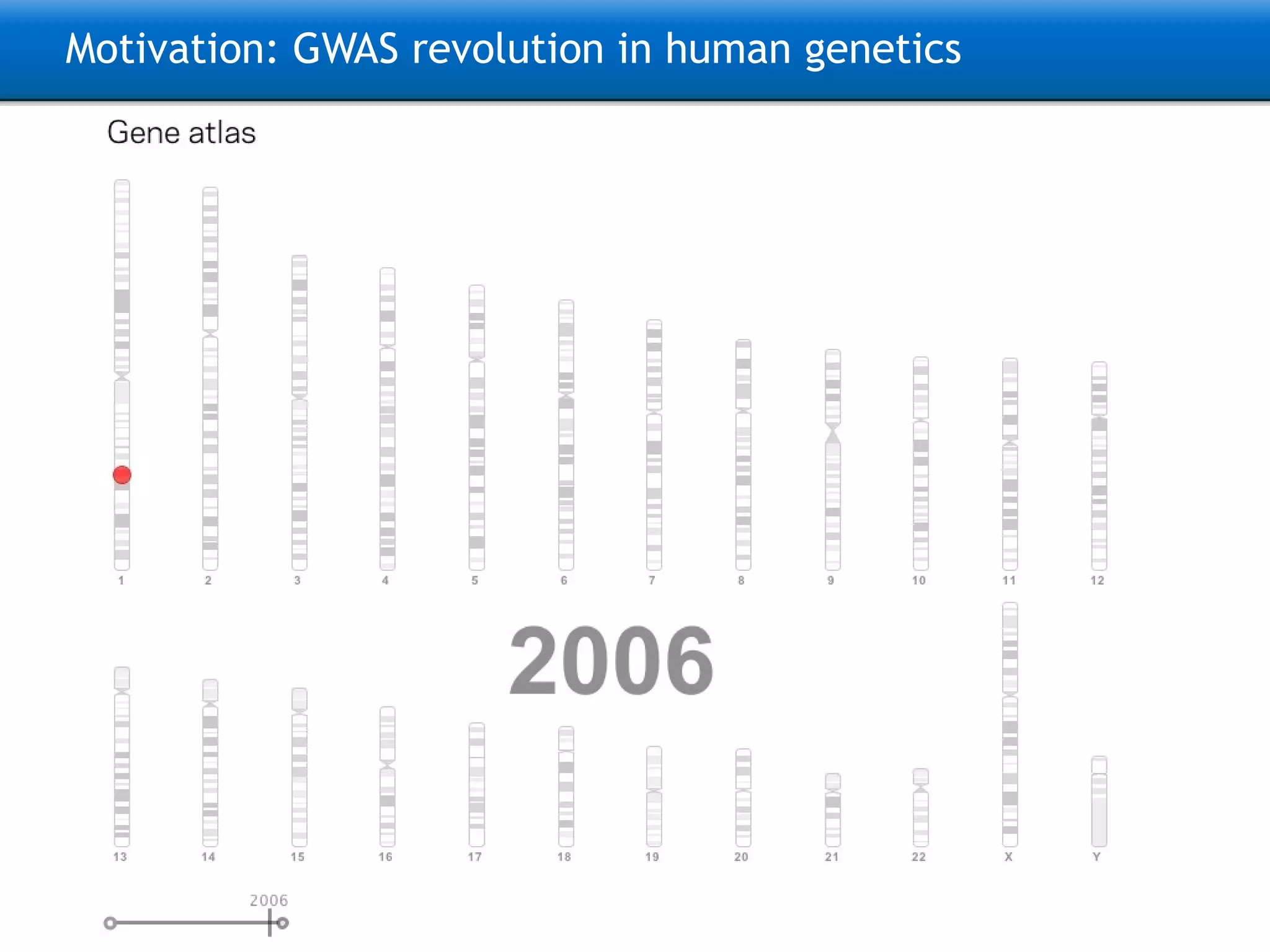
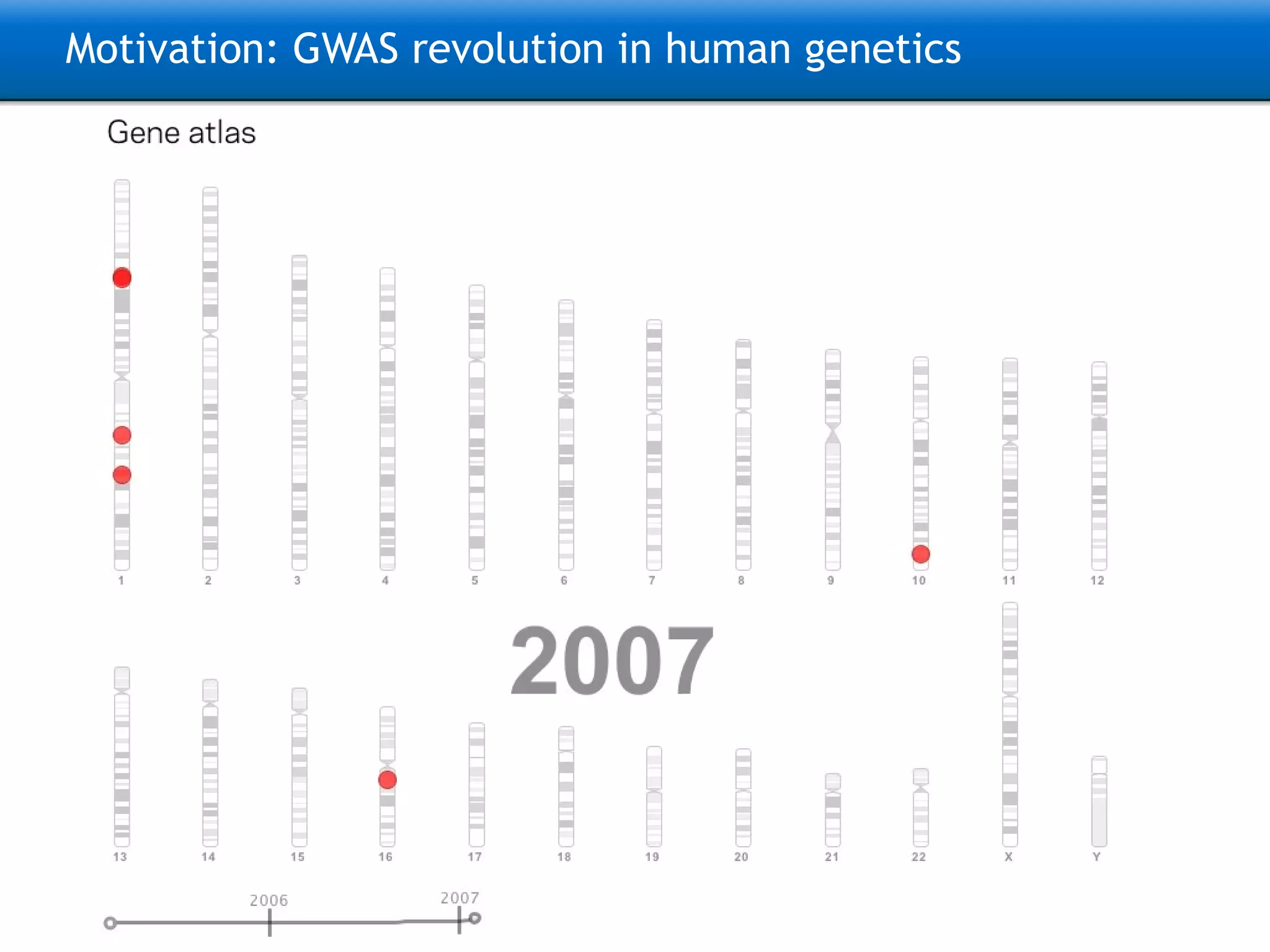
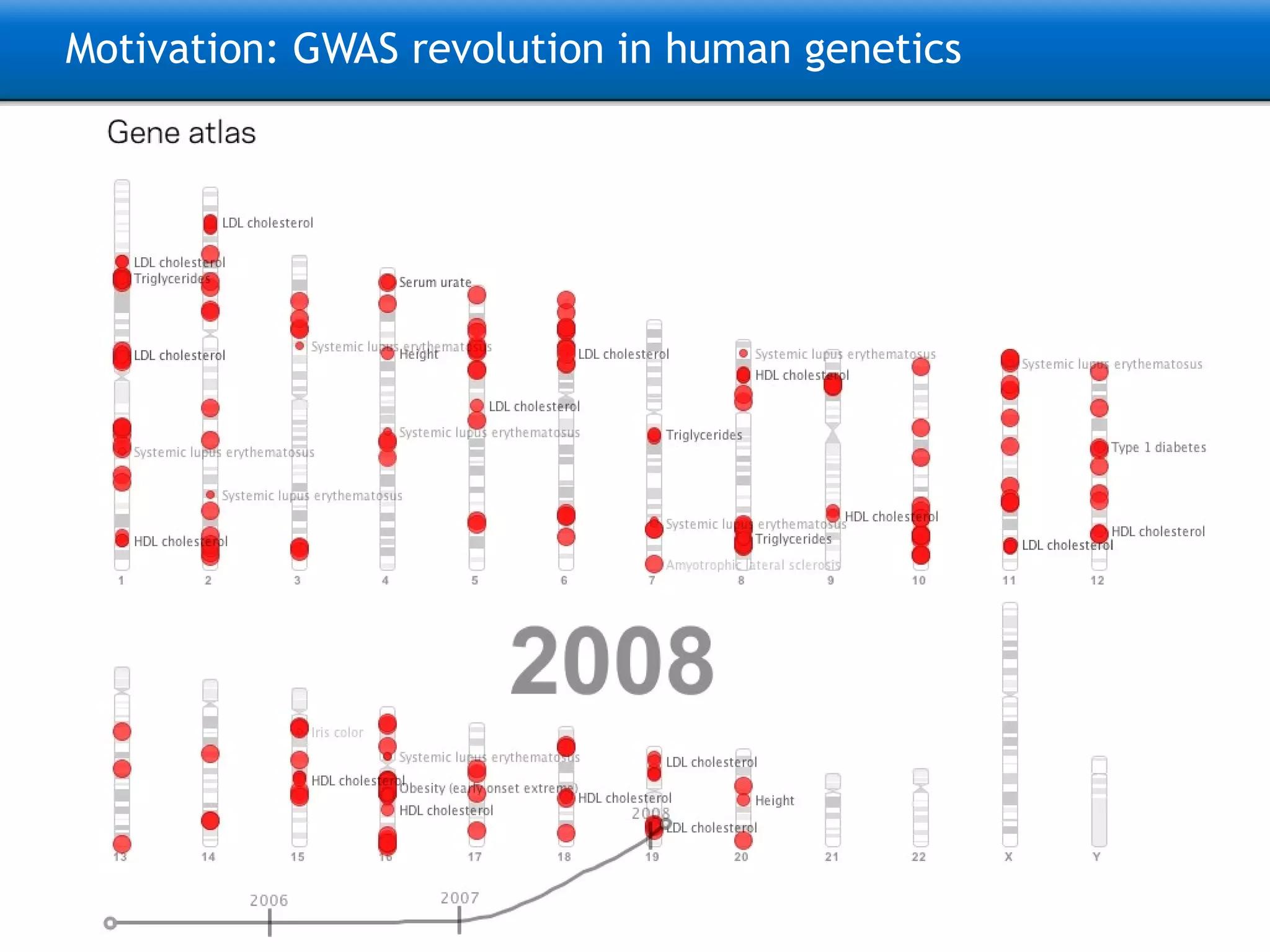
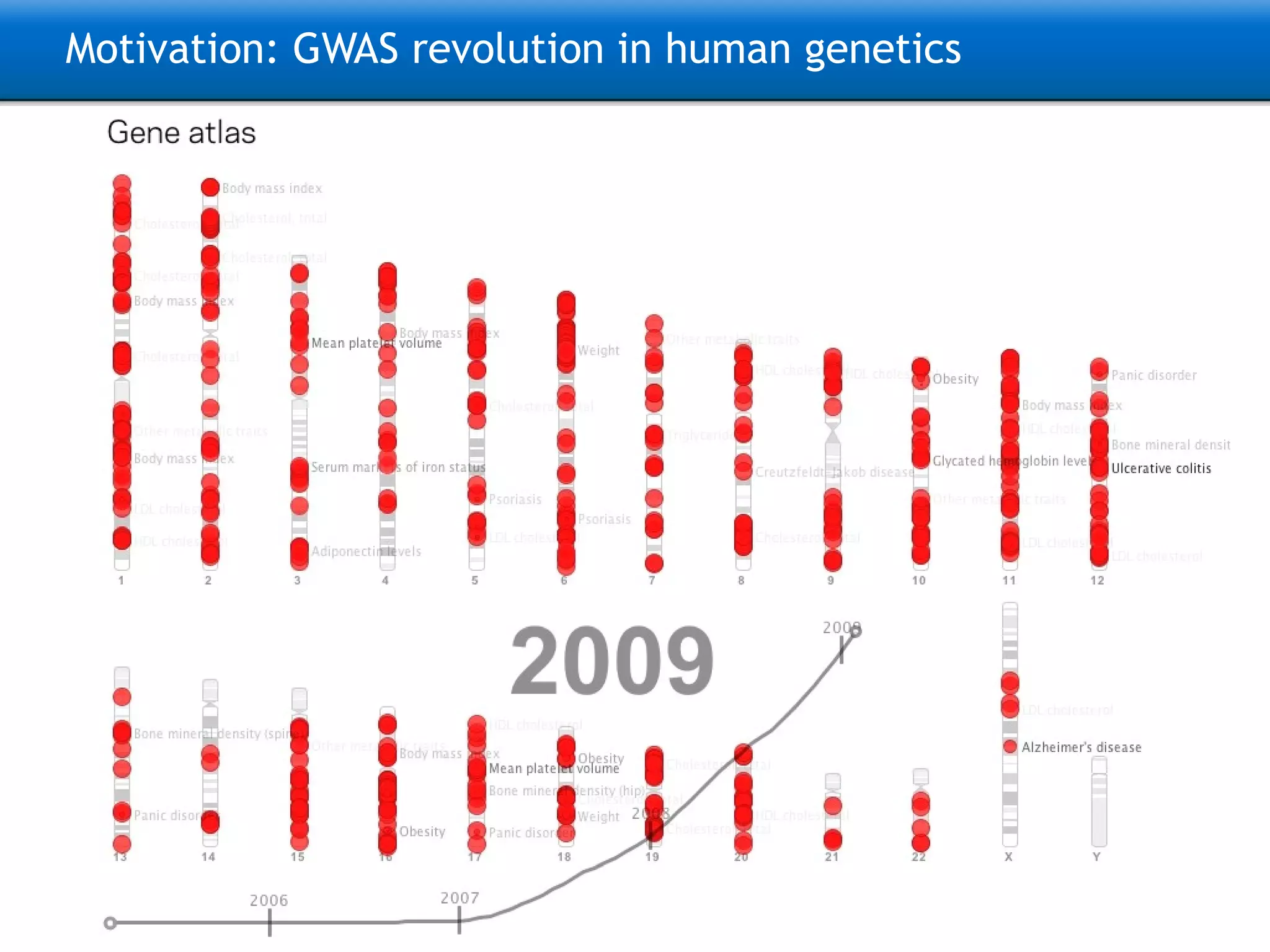
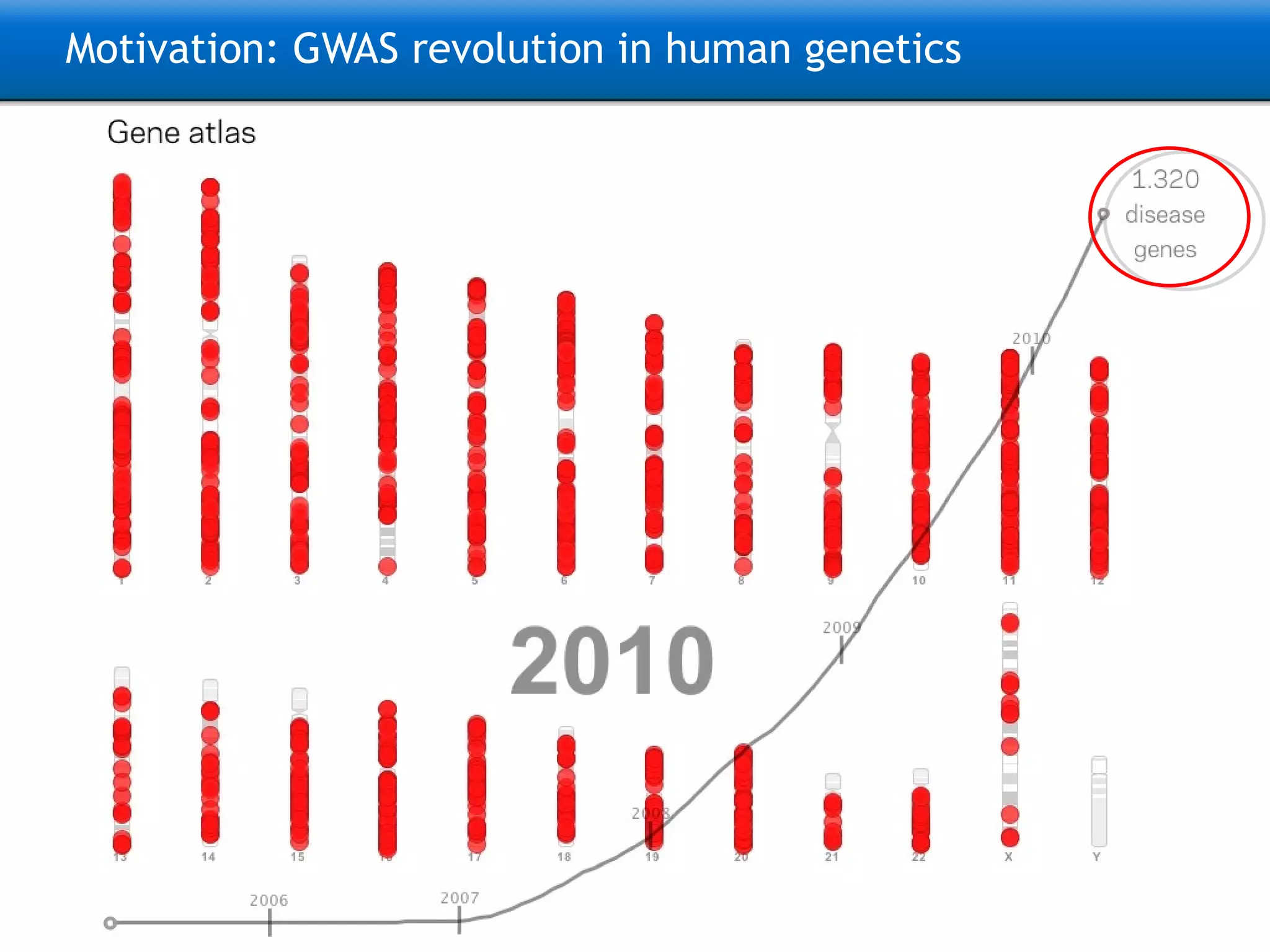
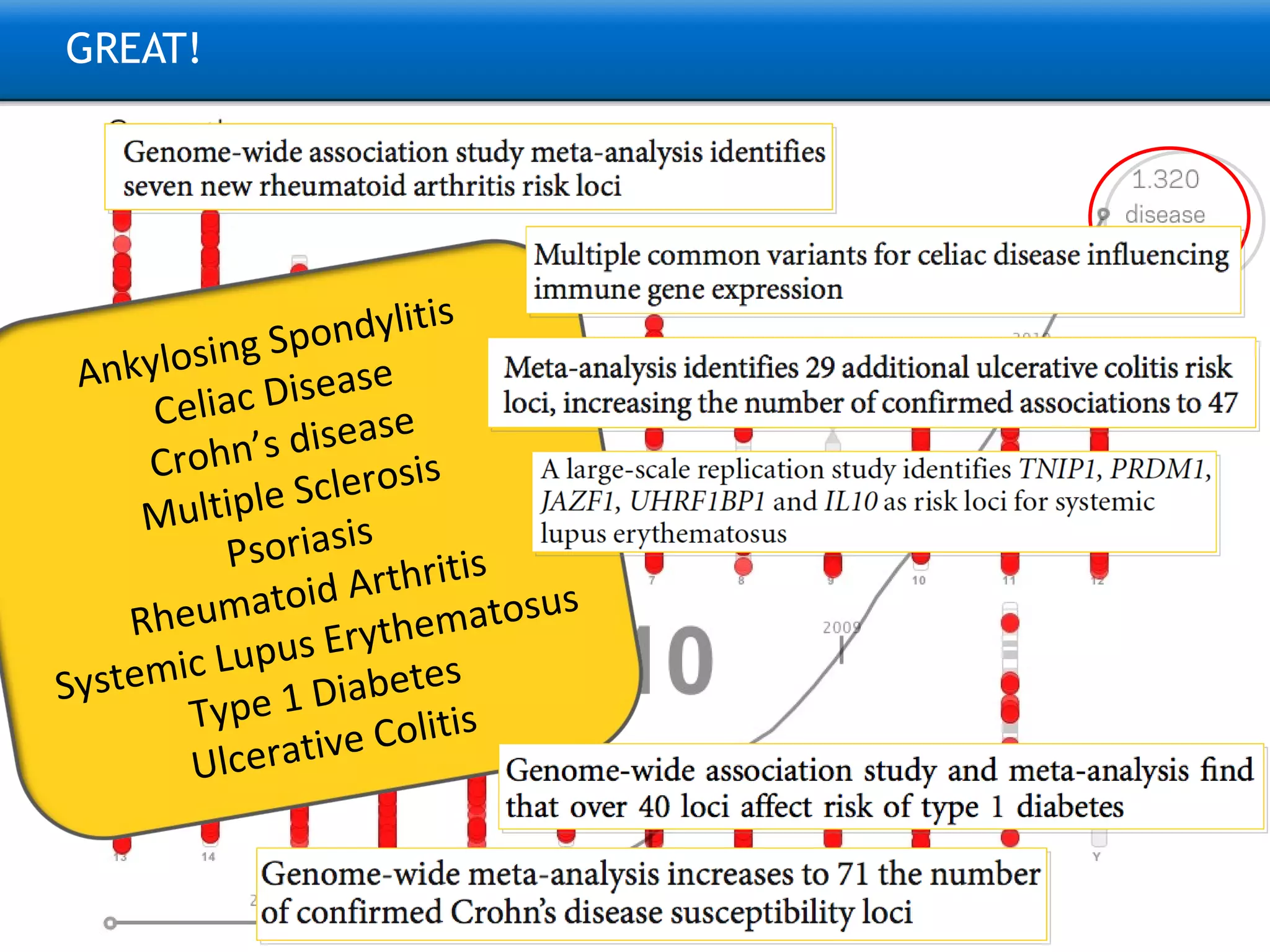
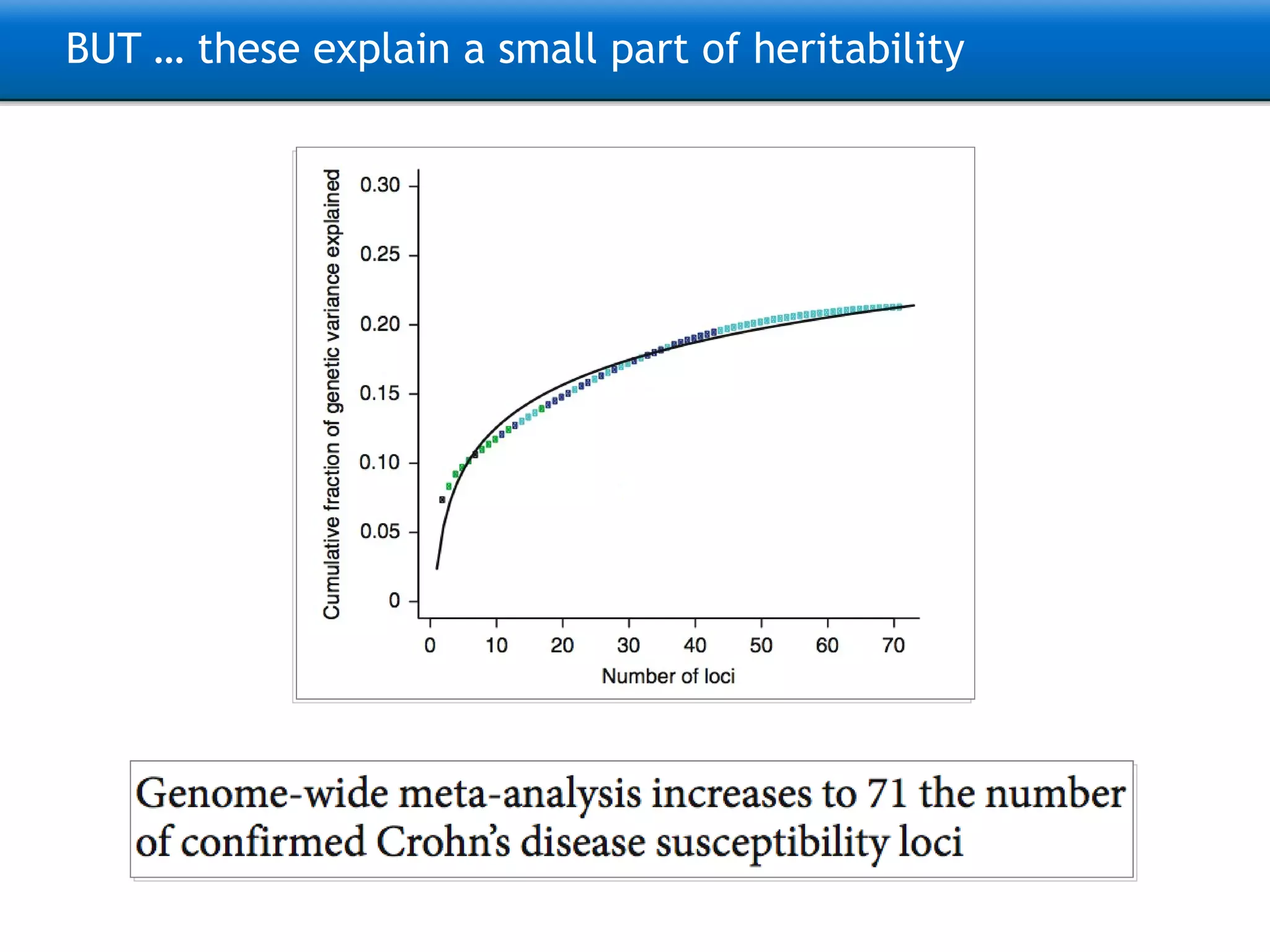
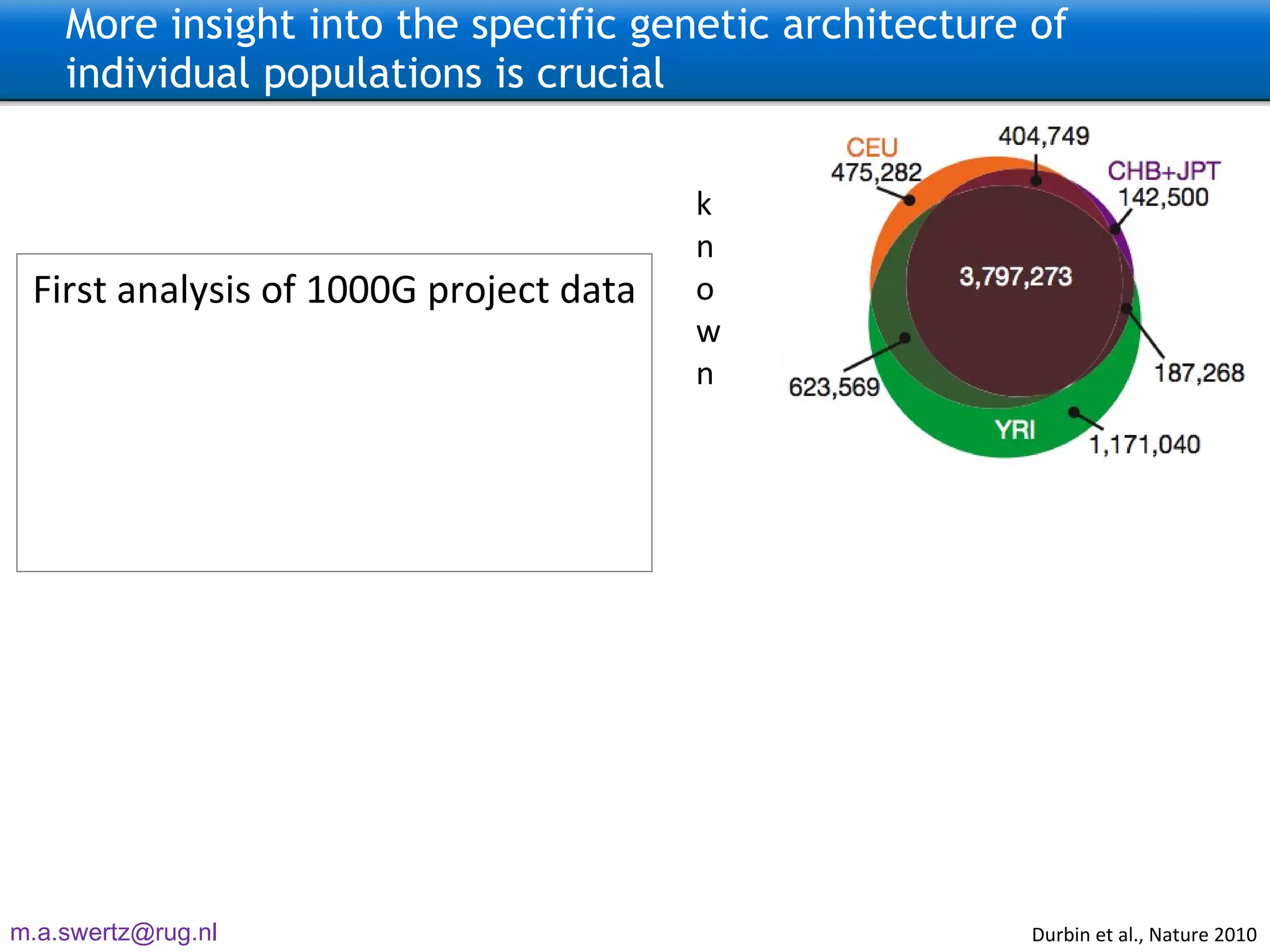
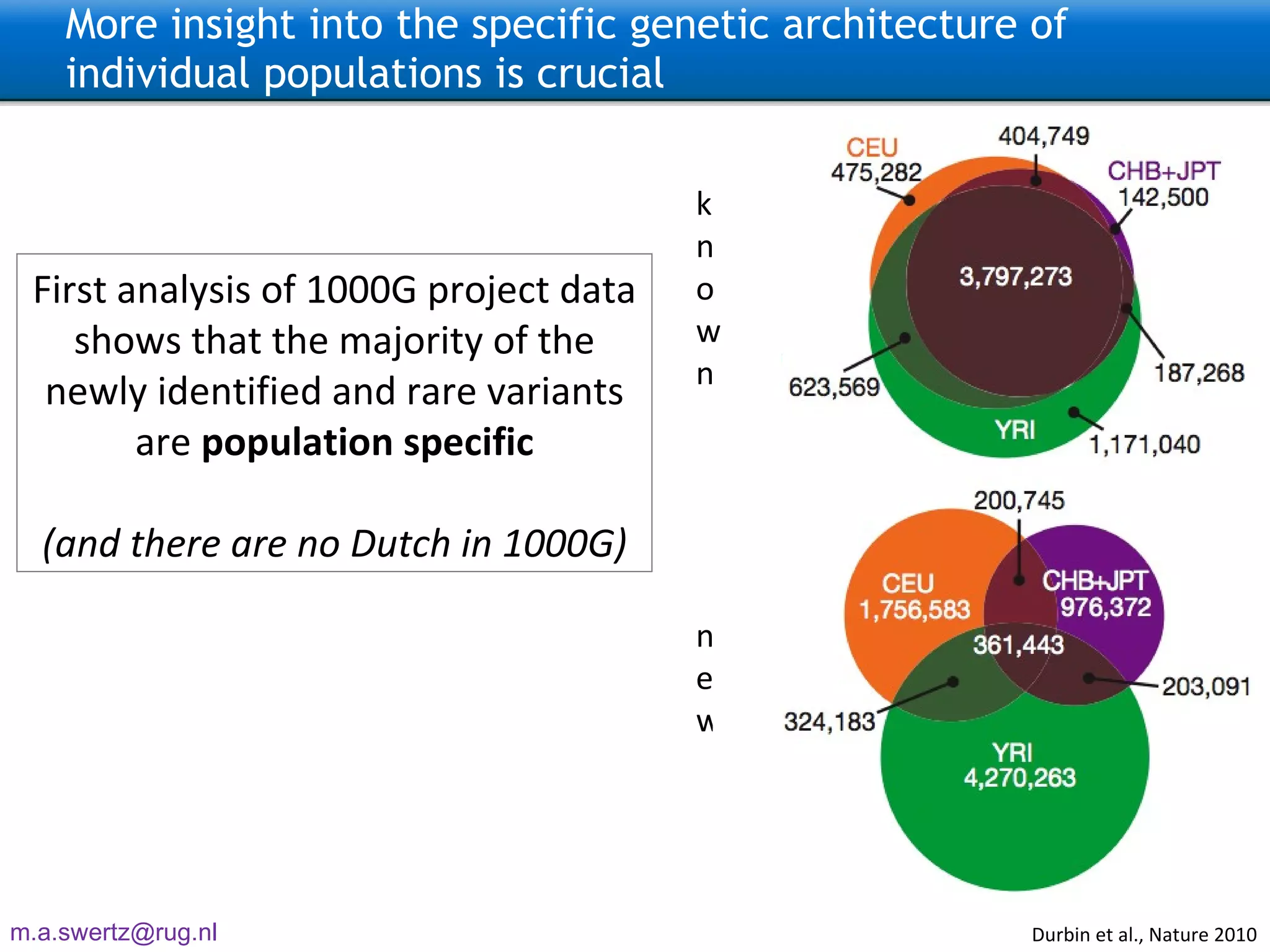
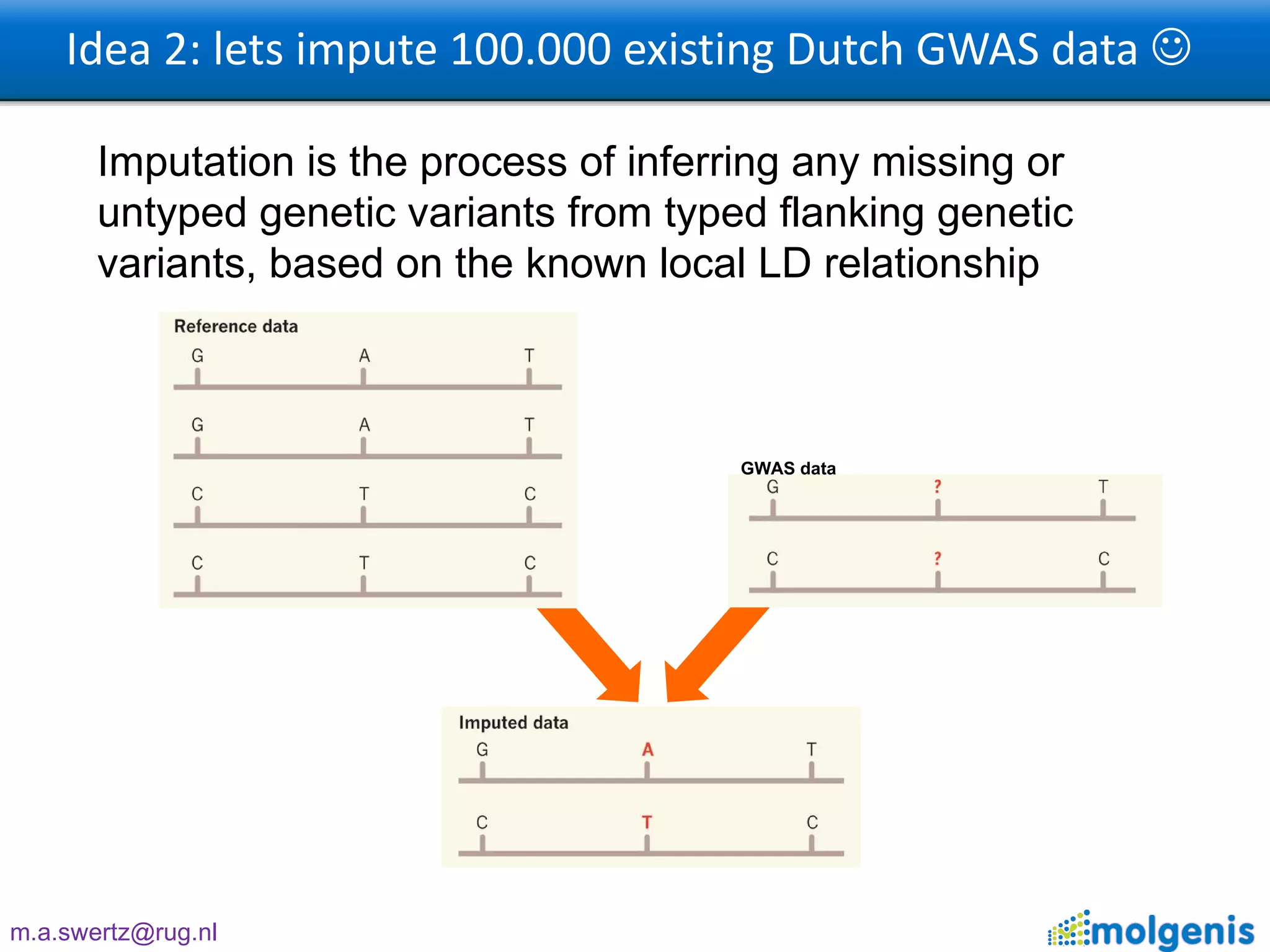
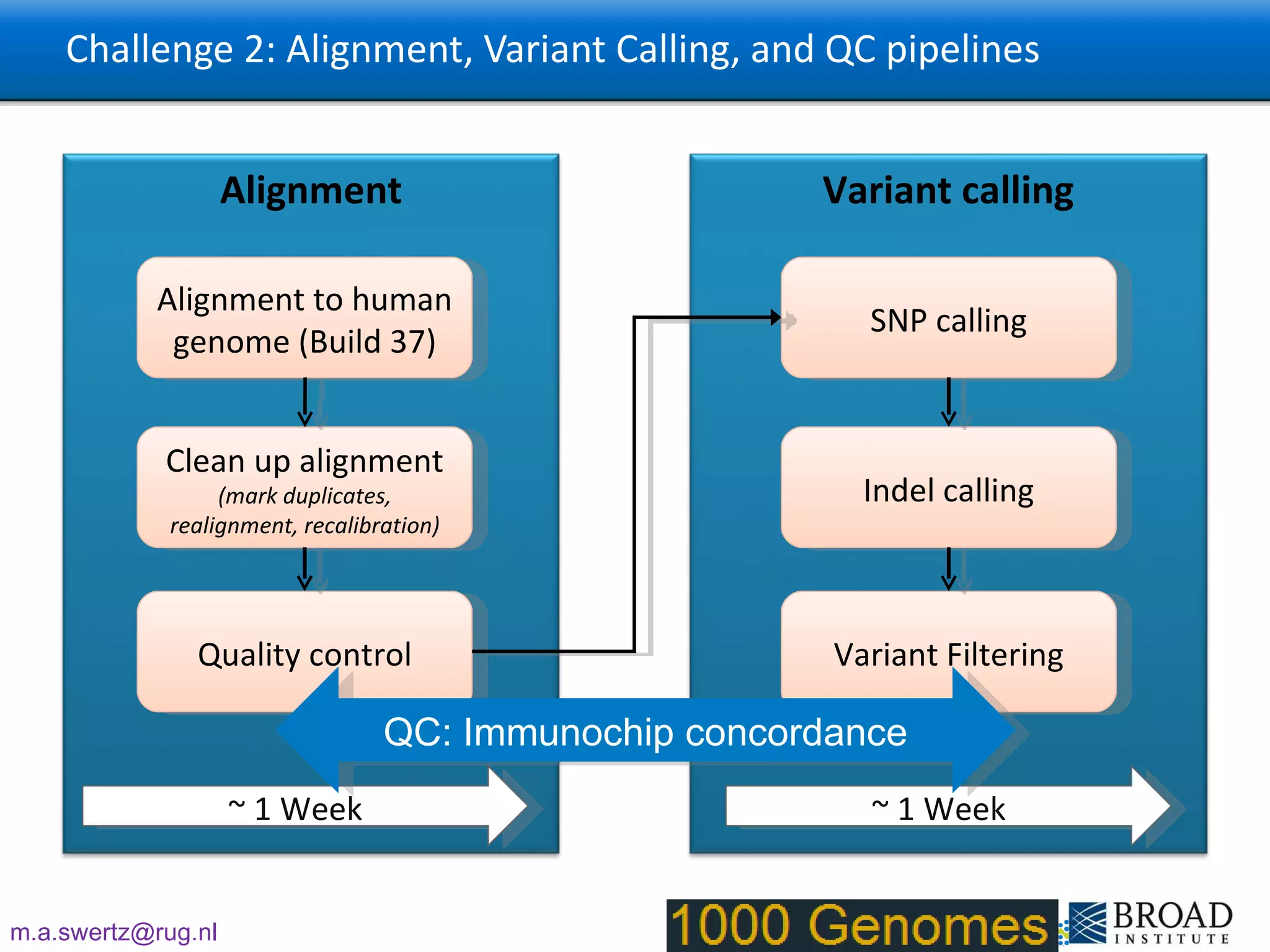
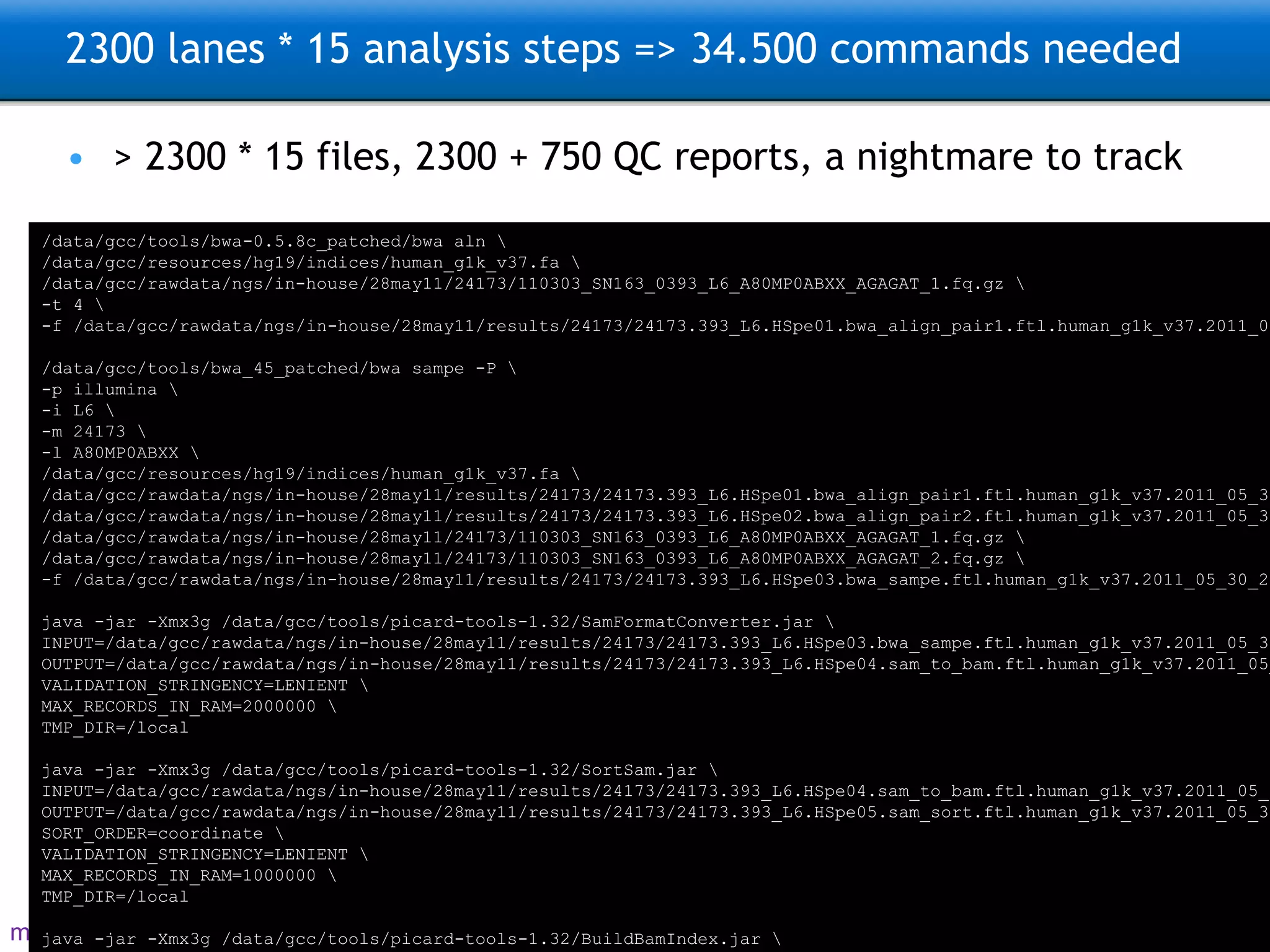
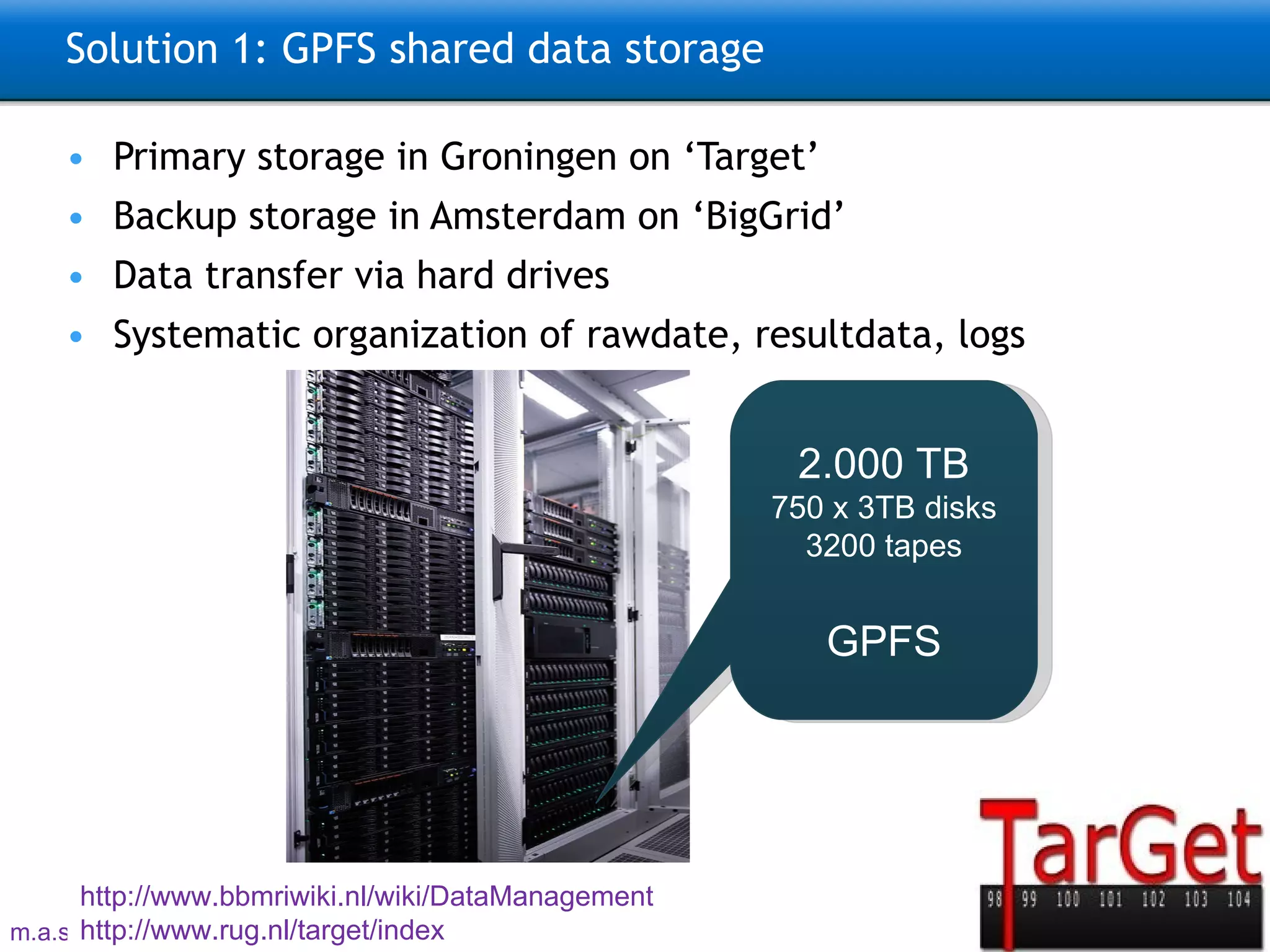
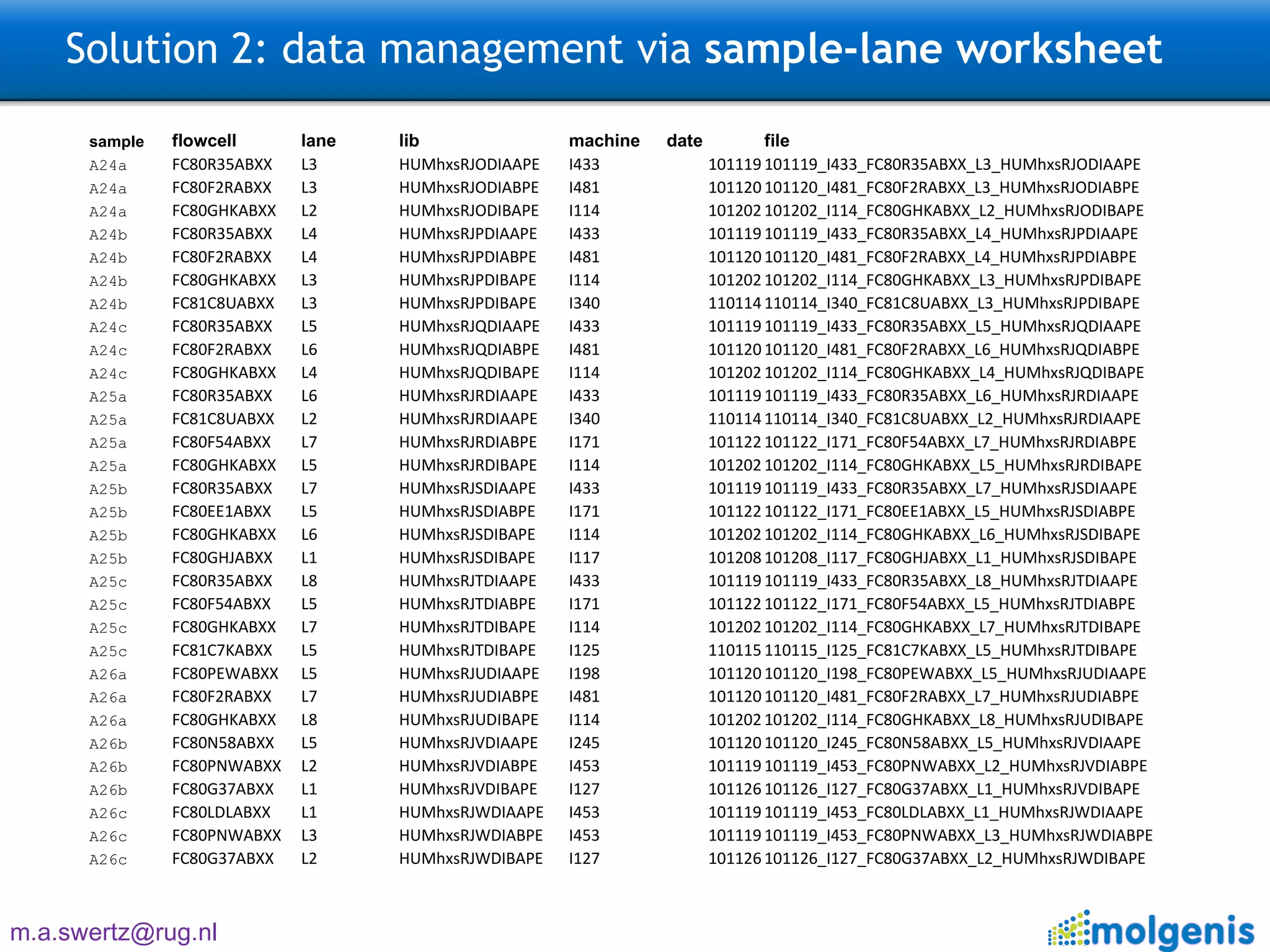
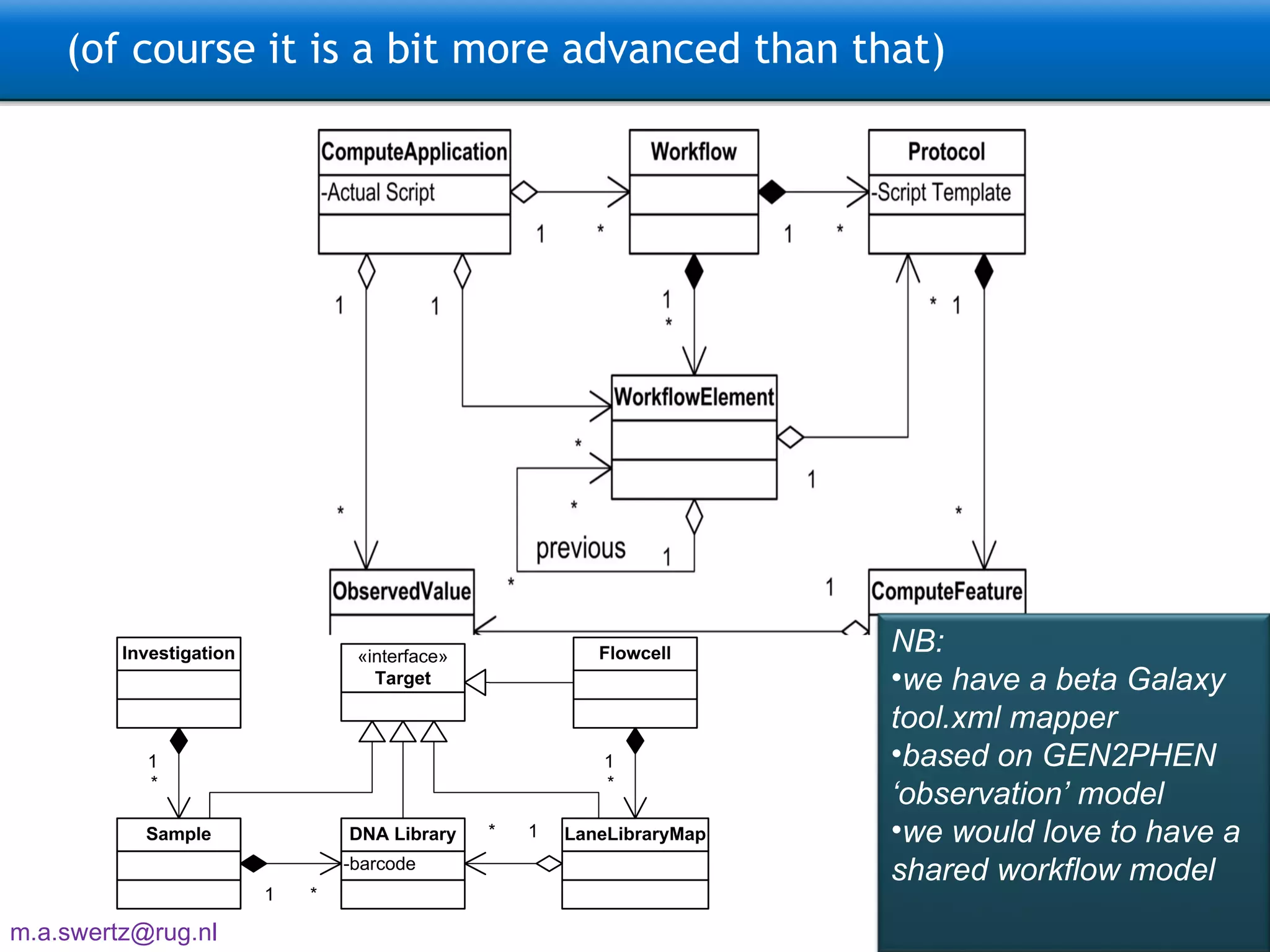
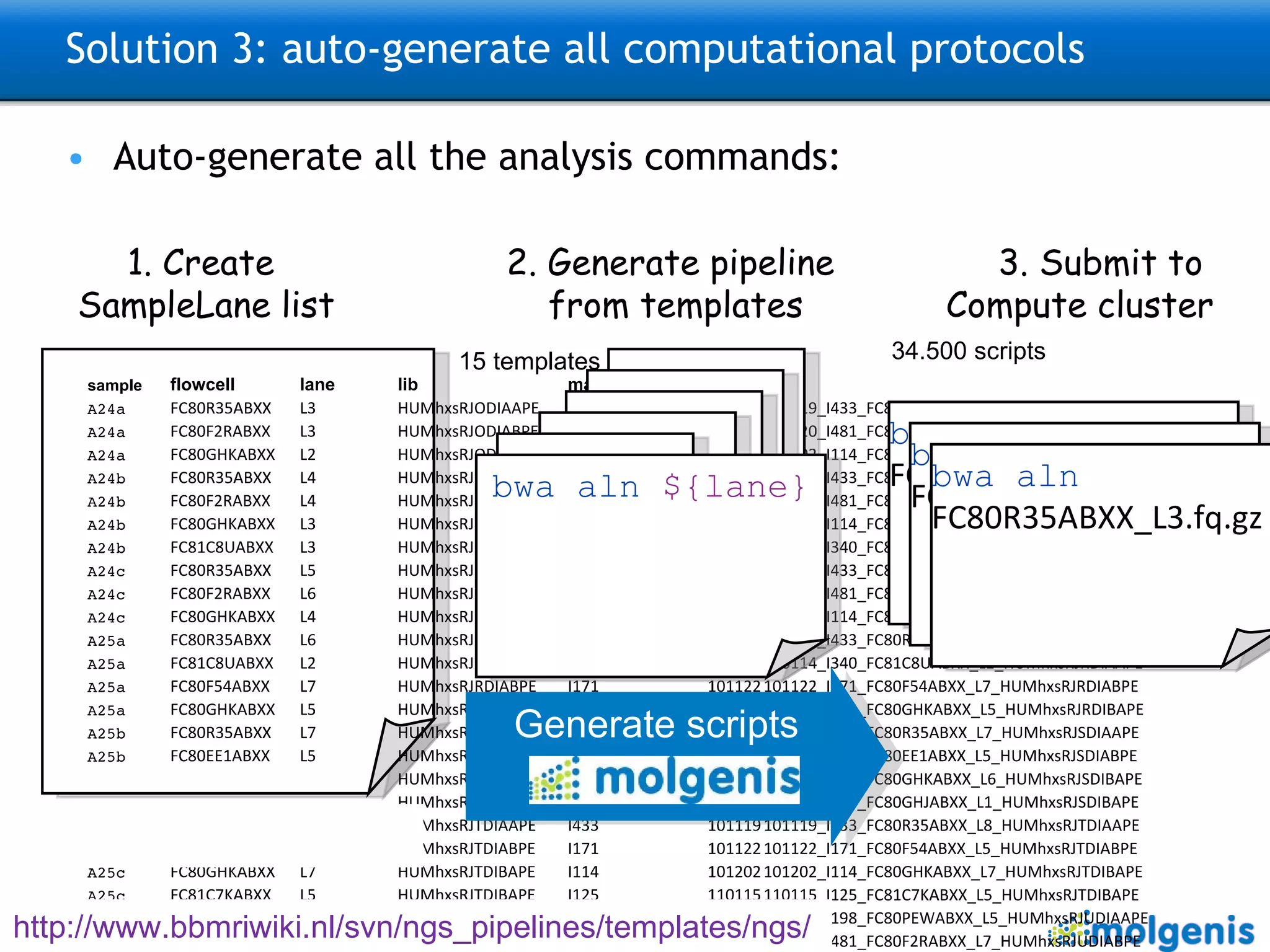
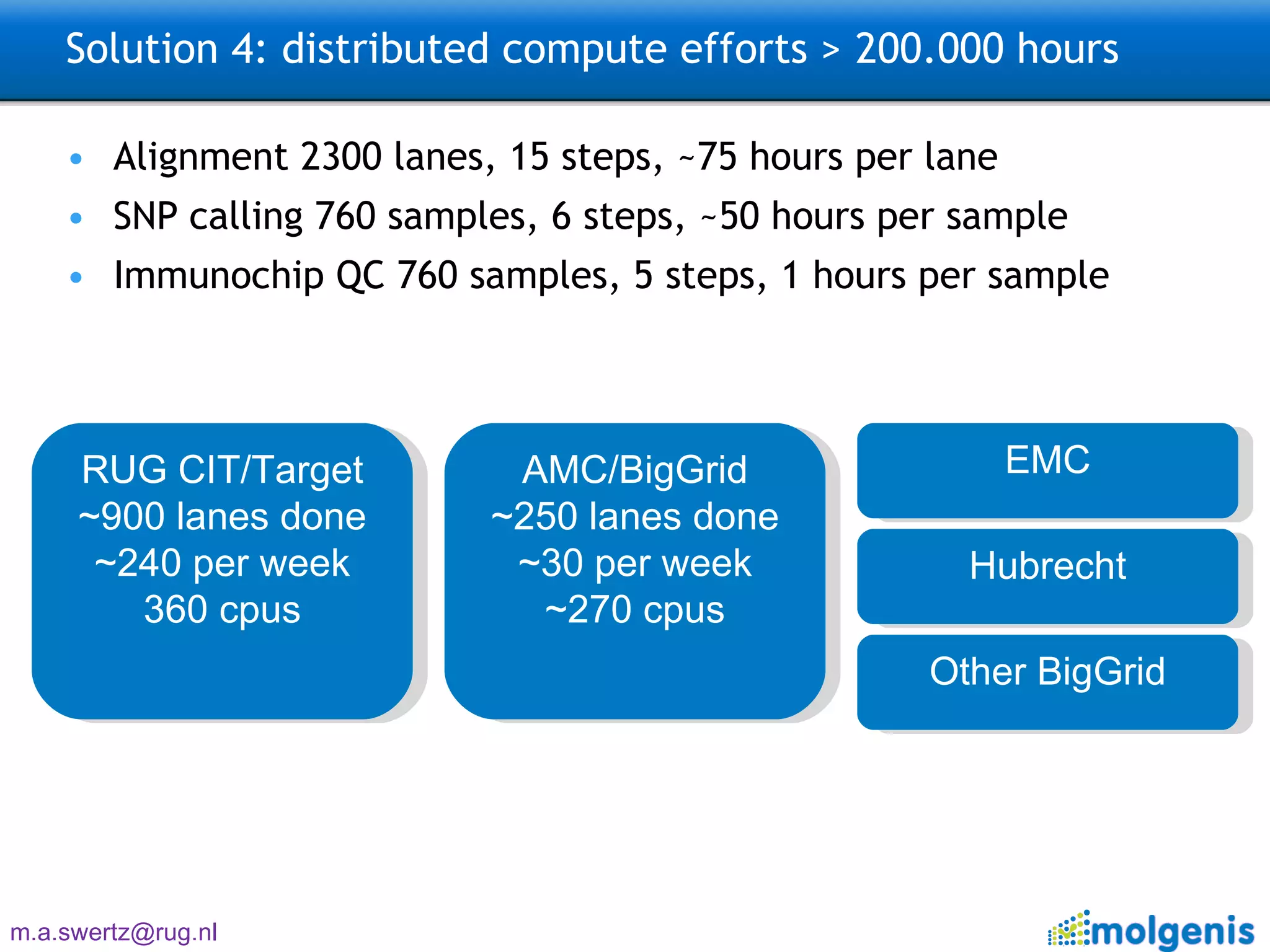
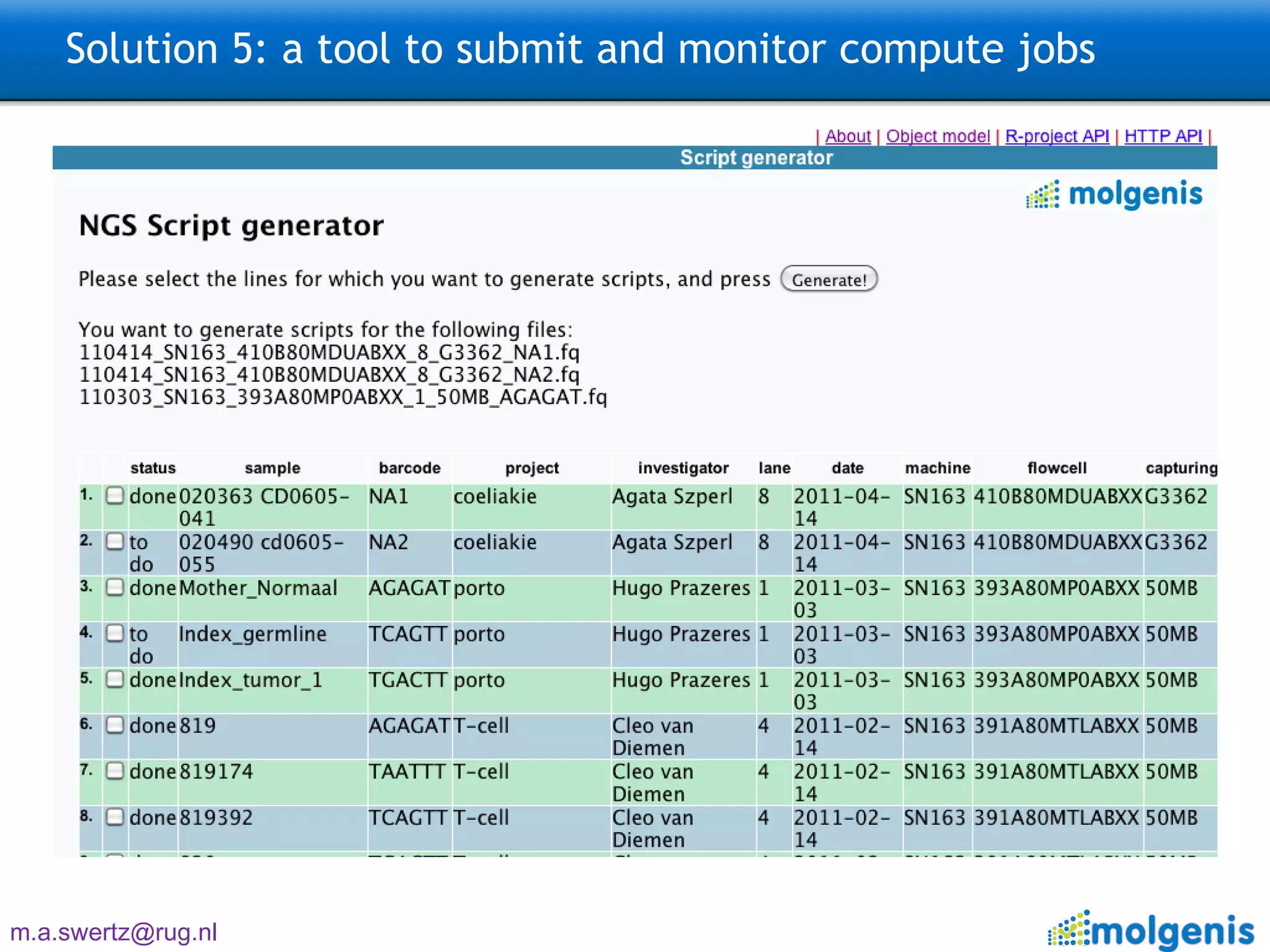
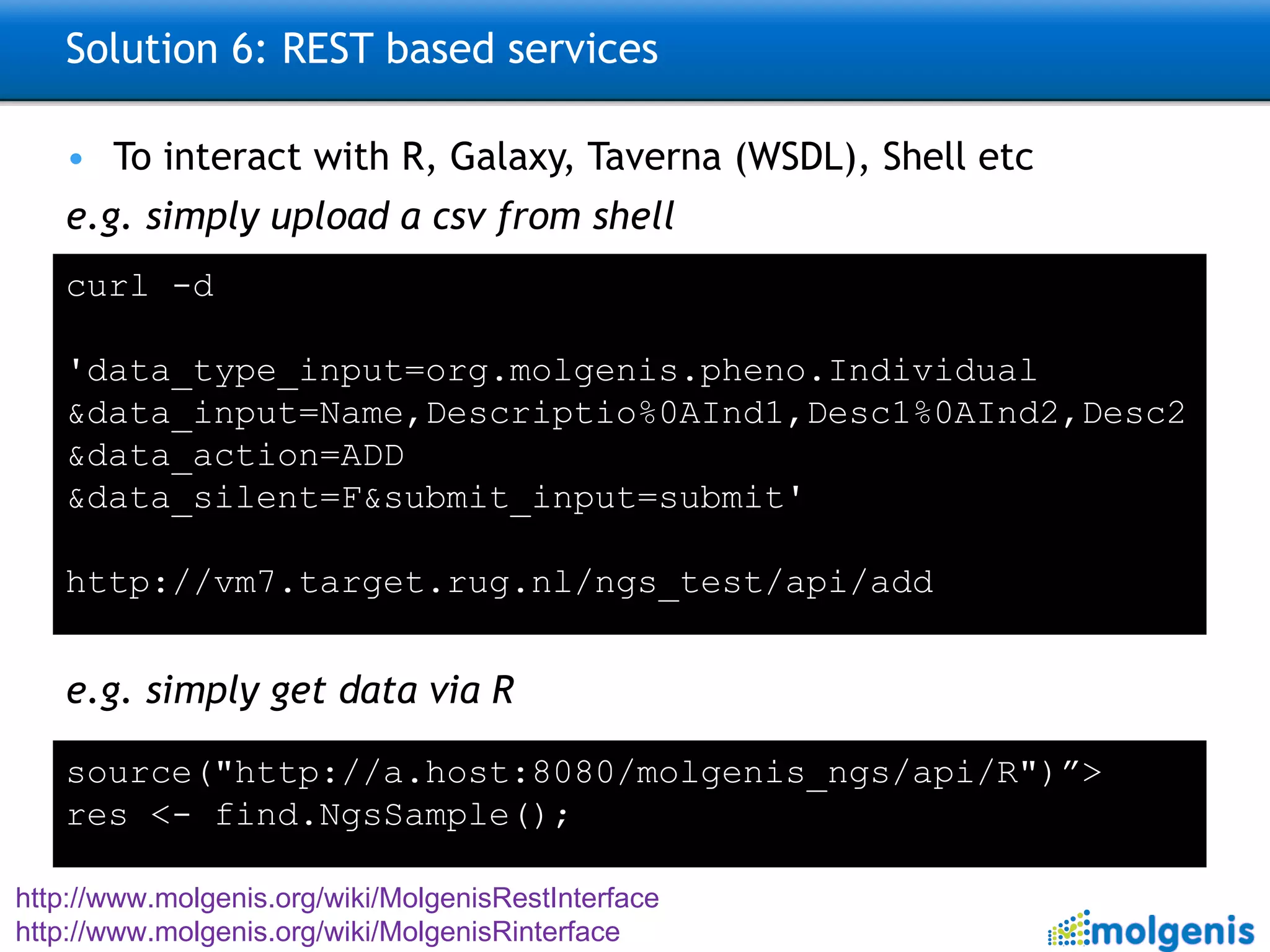
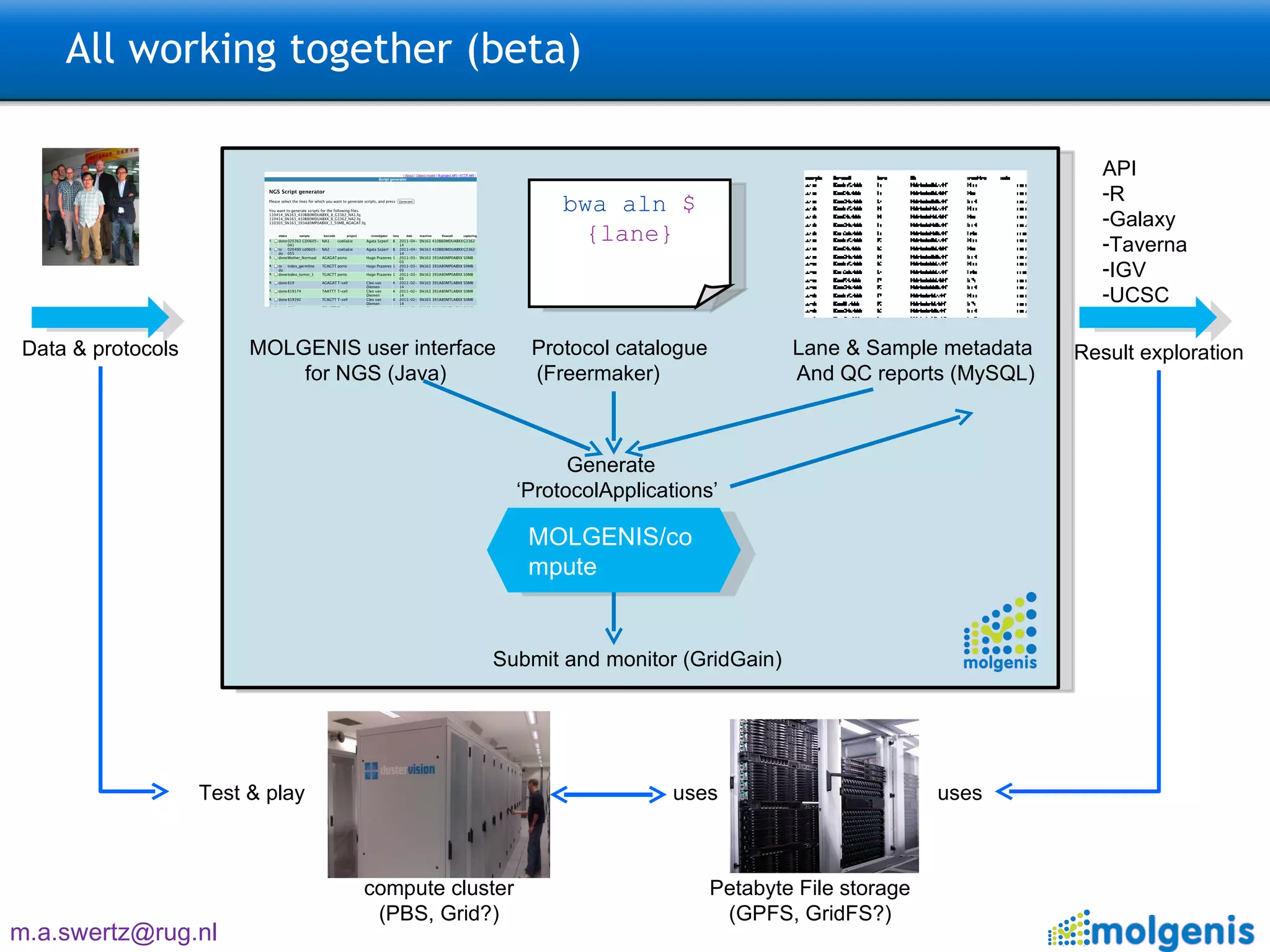
The document describes the implementation of large-scale Next-Generation Sequencing (NGS) pipelines using the Molgenis platform for the Genome of the Netherlands project, aiming to create a Dutch genetic hapmap by sequencing 1000 chromosomes. It outlines challenges faced in data handling and processing, such as alignment and variant calling, and presents the software and hardware infrastructure necessary for these tasks. The project seeks to gain insights into rare genetic variants and improve understanding of the genetic architecture of populations.
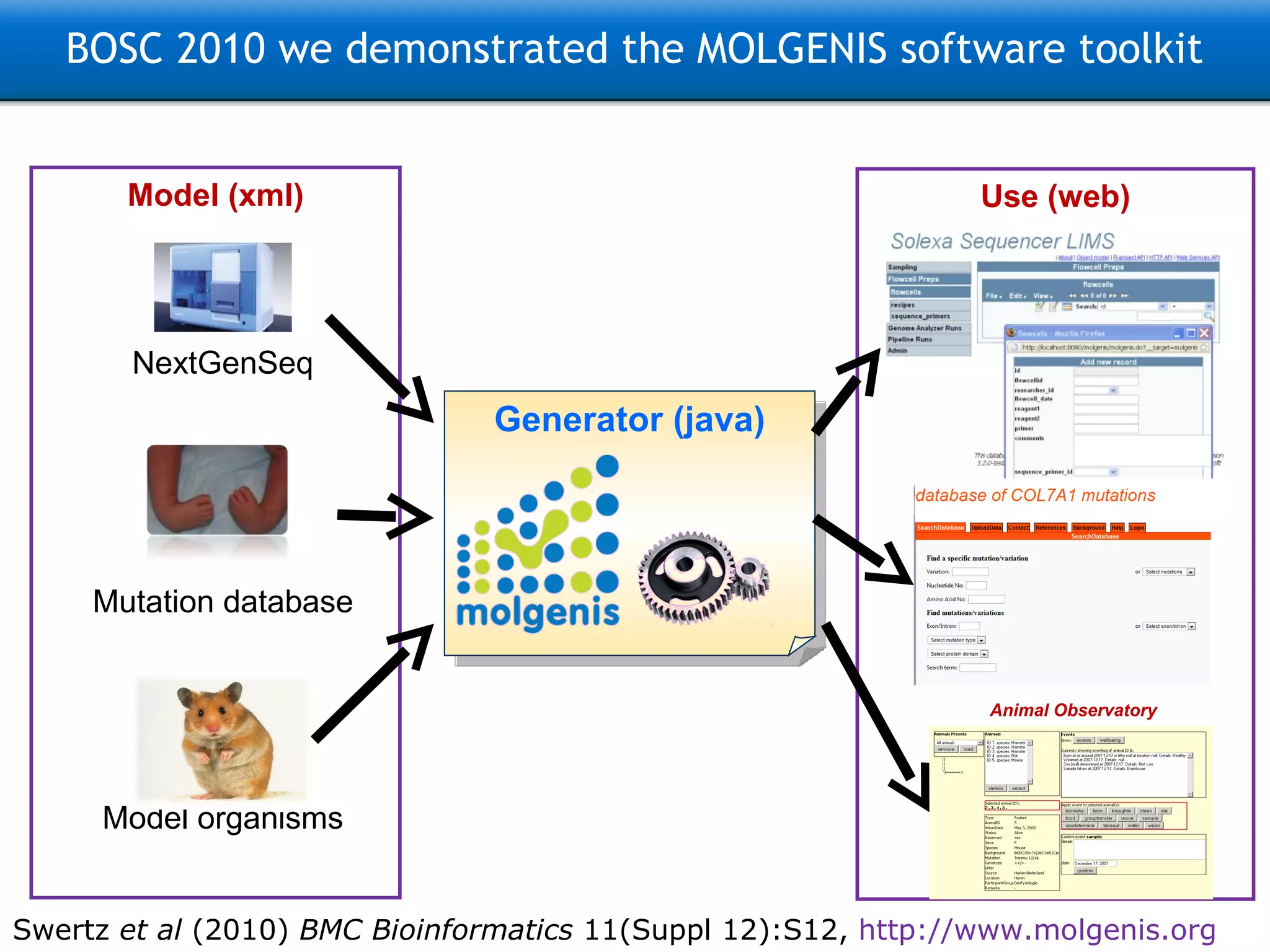
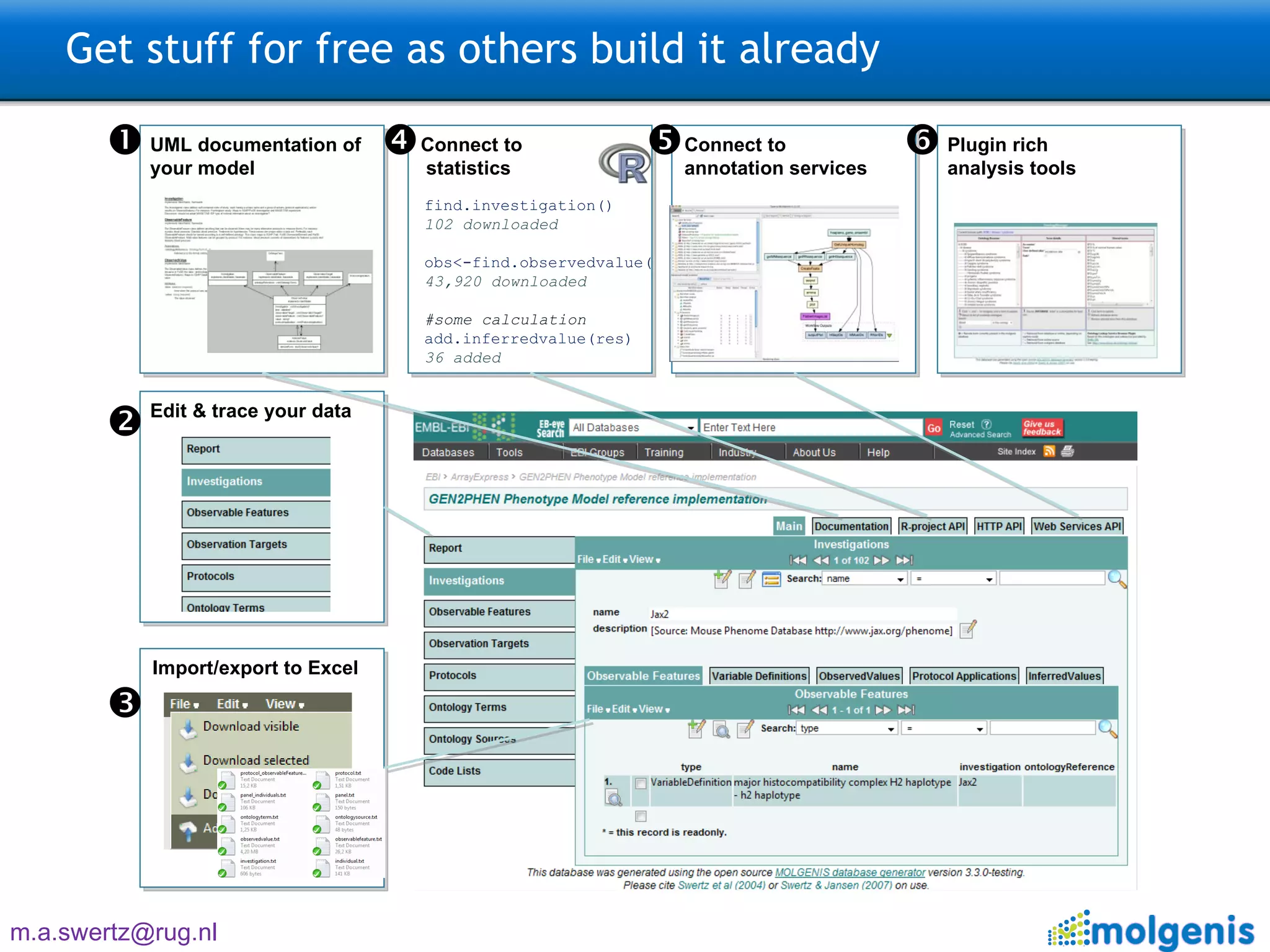
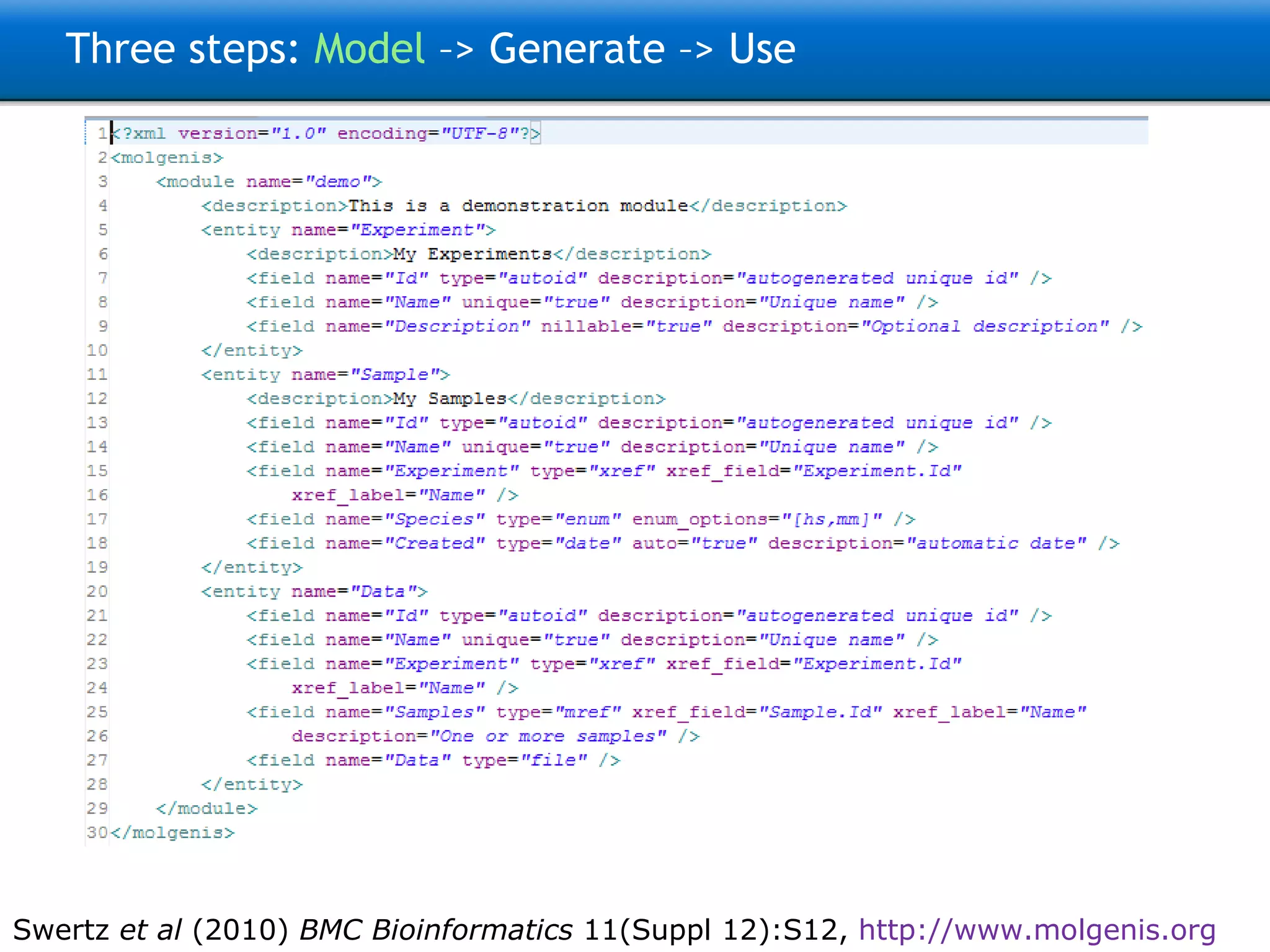
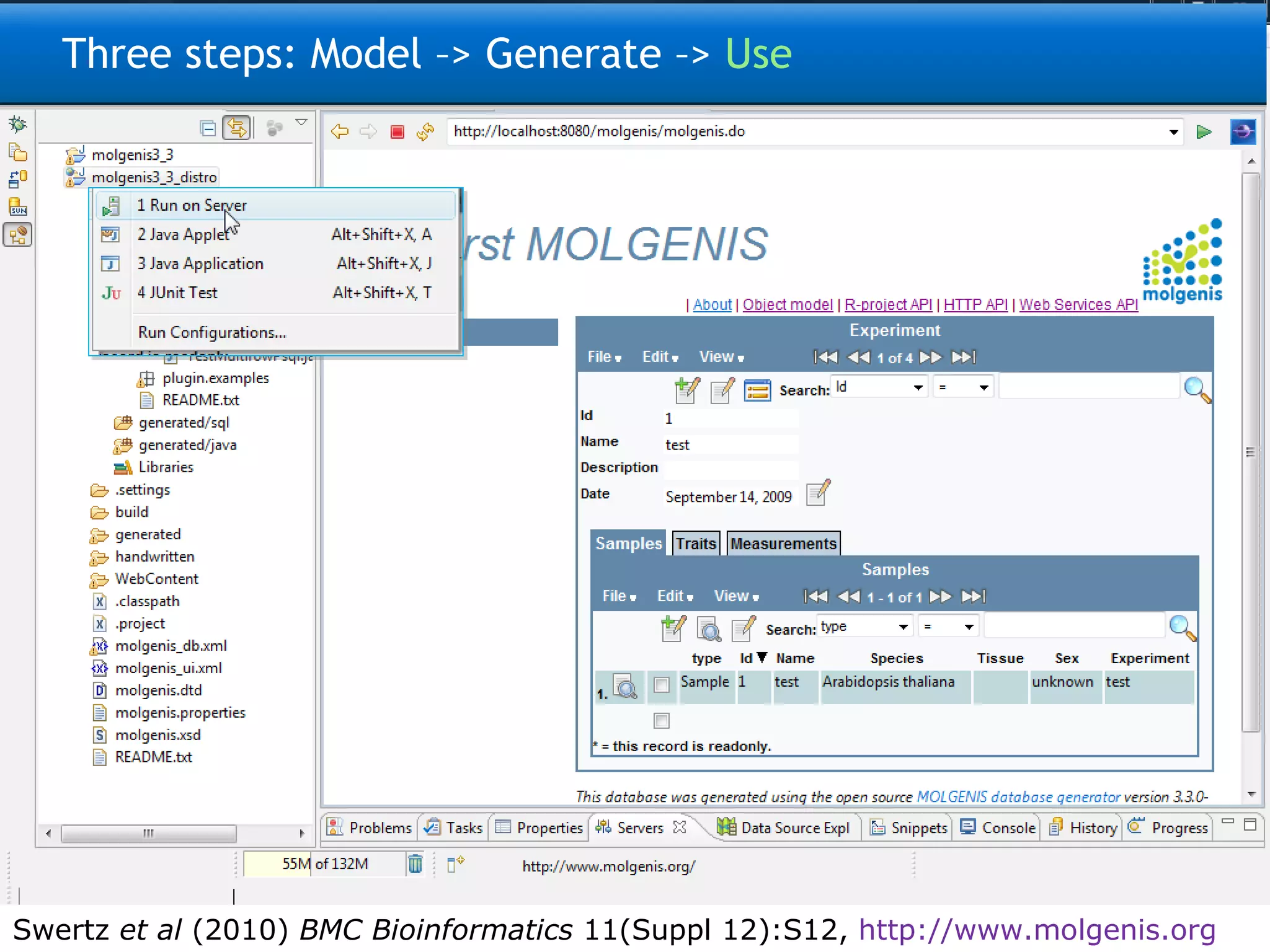
![Three steps: Model –> Generate –> Use 9200 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\ProtocolsForm.java 9293 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\Protocols\ProtocolMenu\ParametersForm.java 9325 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\Protocols\ProtocolMenu\ProtocolComponentsForm.java 9496 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\Ontologies\OntologyTermsForm.java 9528 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\Ontologies\OntologySourcesForm.java 9606 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\Ontologies\OntologySources\OntologyTermsForm.java 9638 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\Ontologies\CodeListsForm.java 9700 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\Ontologies\CodeLists\CodesForm.java 9965 INFO [MenuScreenGen] generated generated\java\ui\screen\TopMenuMenu.java 10012 INFO [MenuScreenGen] generated generated\java\ui\screen\TopMenu\MainMenu.java 10059 INFO [MenuScreenGen] generated generated\java\ui\screen\TopMenu\Main\Investigations\InvestigationMenuMenu.java 10152 INFO [MenuScreenGen] generated generated\java\ui\screen\TopMenu\Main\Investigations\InvestigationMenu\ProtocolApplications\ProtocolApplicationMenuMenu.java 10230 INFO [MenuScreenGen] generated generated\java\ui\screen\TopMenu\Main\ObservationTargetsMenu.java 10293 INFO [MenuScreenGen] generated generated\java\ui\screen\TopMenu\Main\Protocols\ProtocolMenuMenu.java 10324 INFO [MenuScreenGen] generated generated\java\ui\screen\TopMenu\Main\OntologiesMenu.java 11354 INFO [PluginScreenGen] generated Molgenis33Workspace\molgenis4phenotype\generated\java\ui\screen\TopMenu\Main\ReportPlugin.java 11557 INFO [PluginScreenGen] generated Molgenis33Workspace\molgenis4phenotype\generated\java\ui\screen\TopMenu\Main\Ontologies\OntologyManagerPlugin.java 11604 INFO [PluginScreenGen] generated Molgenis33Workspace\molgenis4phenotype\generated\java\ui\screen\TopMenu\Model_documentationPlugin.java 11604 INFO [PluginScreenGen] generated Molgenis33Workspace\molgenis4phenotype\generated\java\ui\screen\TopMenu\RprojectApiPlugin.java 11620 INFO [PluginScreenGen] generated Molgenis33Workspace\molgenis4phenotype\generated\java\ui\screen\TopMenu\HttpApiPlugin.java 11635 INFO [PluginScreenGen] generated Molgenis33Workspace\molgenis4phenotype\generated\java\ui\screen\TopMenu\WebServicesApiPlugin.java 11651 WARN [PluginScreenFTLTemplateGen] Skipped because exists: handwritten\java\plugin\report\InvestigationOverview.ftl 11807 WARN [PluginScreenFTLTemplateGen] Skipped because exists: handwritten\java\plugin\OntologyBrowser\OntologyBrowserPlugin.ftl 11807 WARN [PluginScreenFTLTemplateGen] Skipped because exists: handwritten\java\plugin\topmenu\DocumentationScreen.ftl 11807 WARN [PluginScreenFTLTemplateGen] Skipped because exists: handwritten\java\plugin\topmenu\RprojectApiScreen.ftl 11823 WARN [PluginScreenFTLTemplateGen] Skipped because exists: handwritten\java\plugin\topmenu\HttpAPiScreen.ftl 11823 WARN [PluginScreenFTLTemplateGen] Skipped because exists: handwritten\java\plugin\topmenu\SoapApiScreen.ftl 11854 WARN [PluginScreenJavaTemplateGen] Skipped because exists: handwritten\java\plugin\report\InvestigationOverview.java 12057 WARN [PluginScreenJavaTemplateGen] Skipped because exists: handwritten\java\plugin\OntologyBrowser\OntologyBrowserPlugin.java 12072 WARN [PluginScreenJavaTemplateGen] Skipped because exists: handwritten\java\plugin\topmenu\DocumentationScreen.java 12088 WARN [PluginScreenJavaTemplateGen] Skipped because exists: handwritten\java\plugin\topmenu\RprojectApiScreen.java 12088 WARN [PluginScreenJavaTemplateGen] Skipped because exists: handwritten\java\plugin\topmenu\HttpAPiScreen.java 12088 WARN [PluginScreenJavaTemplateGen] Skipped because exists: handwritten\java\plugin\topmenu\SoapApiScreen.java 12103 INFO [MolgenisServletContextGen] generated WebContent\META-INF\context.xml 12259 INFO [SoapApiGen] generated generated\java\ui\SoapApi.java 12353 INFO [CsvExportGen] generated generated\java\tools\CsvExport.java 12431 INFO [CsvImportByNameGen] generated generated\java\tools\CsvImportByName.java 12636 INFO [CopyMemoryToDatabaseGen] generated generated\java\ui\tools\CopyMemoryToDatabase.java Real example: Generates 150 files, 30k lines of Java, MySQL, CXF, Tomcat config, and R code + docs](https://image.slidesharecdn.com/swertzbosc2011-110718162003-phpapp01/75/D02-NextGenSeq-MOLGENIS-5-2048.jpg)



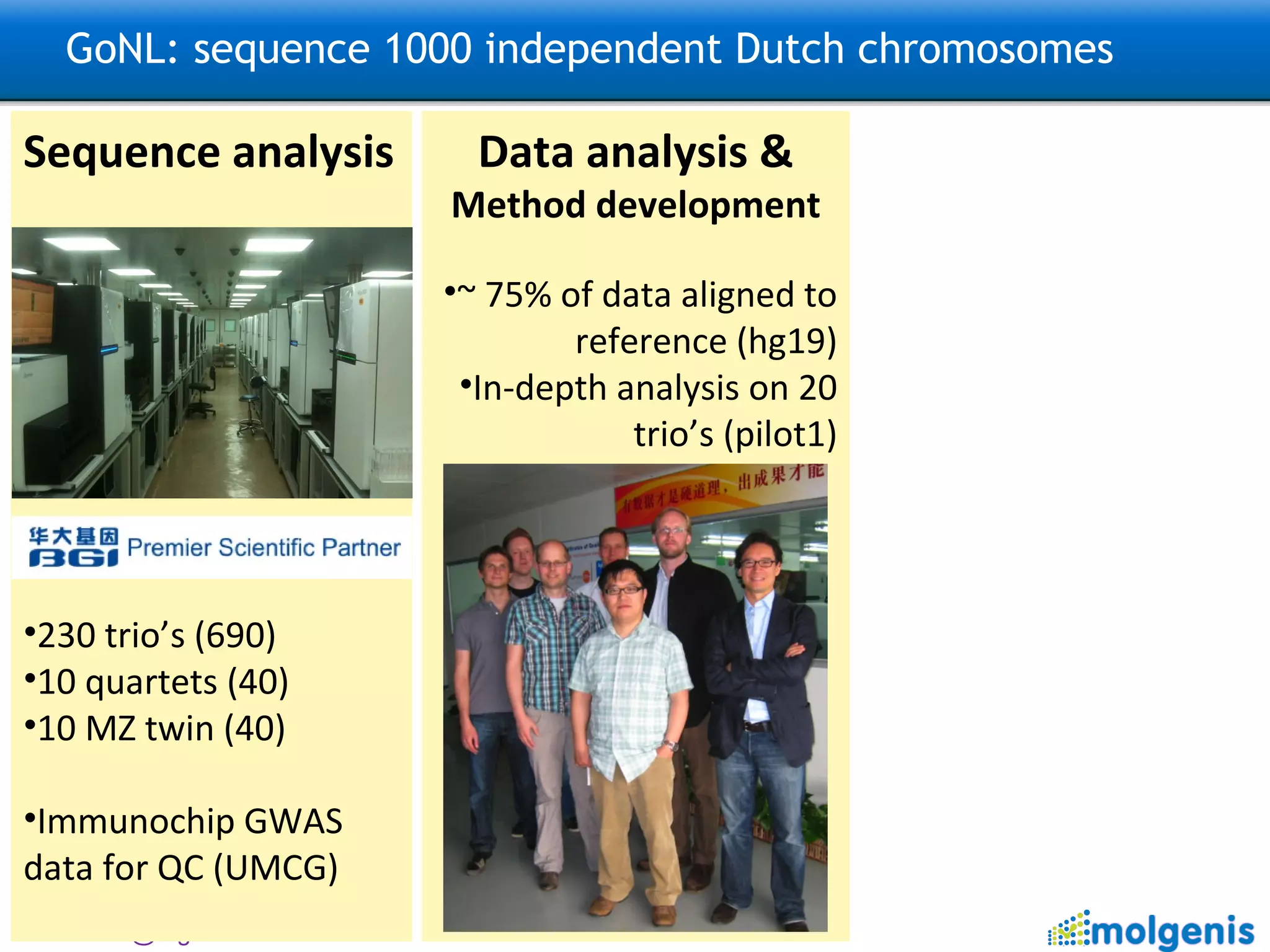
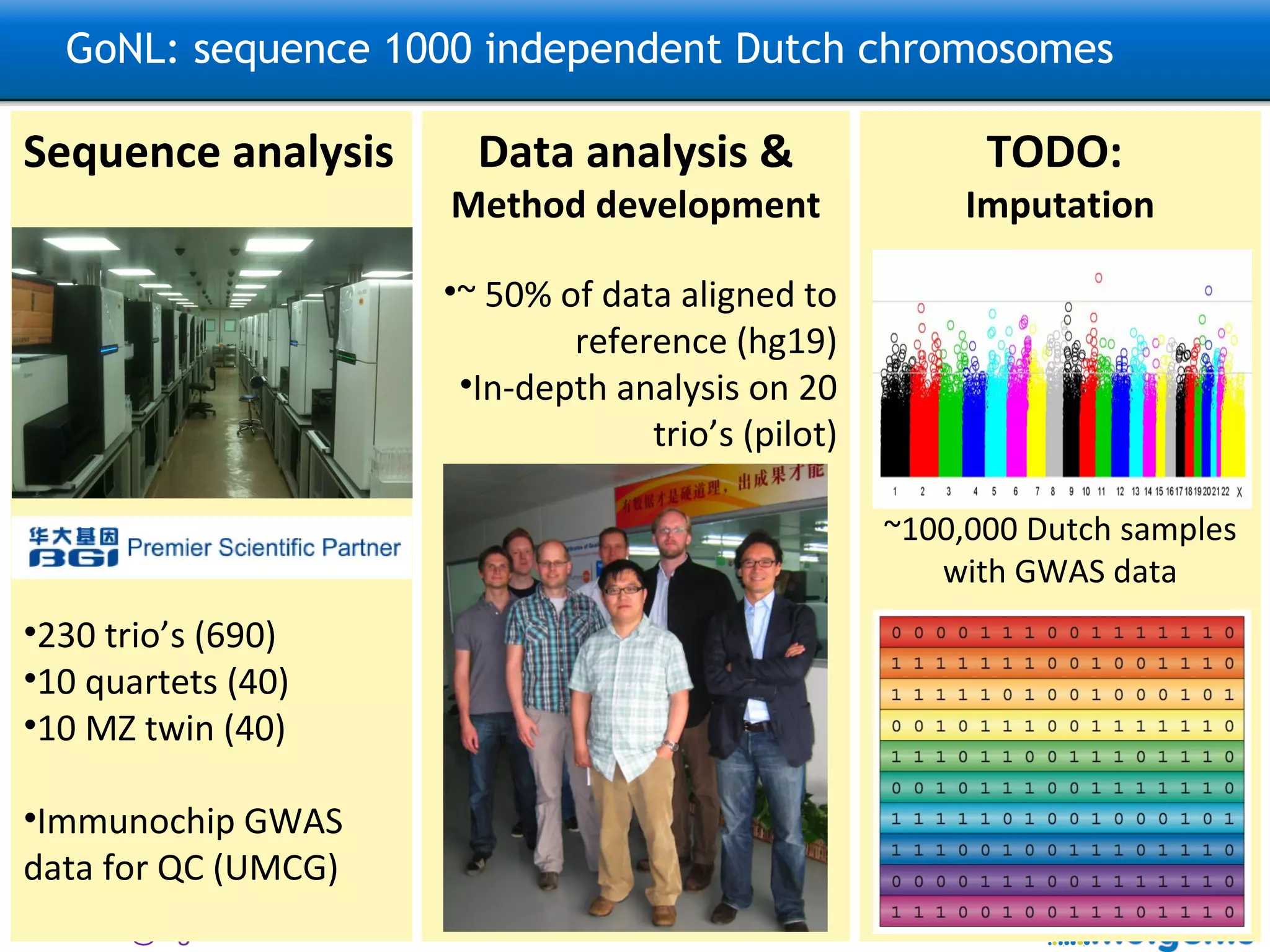
![GoNL: sequence 1000 independent Dutch chromosomes Sequence analysis 230 trio’s (690) 10 quartets (40) 10 MZ twin (40) TODO: Imputation ~100,000 Dutch samples with GWAS data Data analysis & Method development ~ 50% of data aligned to reference (hg19) In-depth analysis on 20 trio’s (pilot) TODO: Further analysis Structural variation, Population Genetics, De novo mutations, Mitochondrial DNA This is an open national project: please contact [email_address] [email_address] and [email_address] for analysis ideas.](https://image.slidesharecdn.com/swertzbosc2011-110718162003-phpapp01/75/D02-NextGenSeq-MOLGENIS-27-2048.jpg)
![GoNL: sequence 1000 independent Dutch chromosomes Data analysis & Method development ~ 75% of data aligned to reference (hg19) In-depth analysis on 20 trio’s (pilot) Sequence analysis 230 trio’s (690) 10 quartets (40) 10 MZ twin (40) Imputation existing GWAS ~100,000 Dutch samples with GWAS data Further analysis Structural variation, Population Genetics, De novo mutations, Mitochondrial DNA This is an open national project: please contact debakker@broadinstitute.org; m.a.swertz@rug.nl; [email_address] for analysis ideas.](https://image.slidesharecdn.com/swertzbosc2011-110718162003-phpapp01/75/D02-NextGenSeq-MOLGENIS-28-2048.jpg)

![Acknowledgements GoNL / MOLGENIS Infrastructure team George Byelas, Martijn Dijkstra, Robert Wagner, Pieter Neerincx, Abhishek Narain, Jan Bot and indirectly GEN2PHEN, EBI, FIMM, ... GoNL Analysis team (creating pipelines and tools) Freerk van Dijk (UMCG), Barbera van Schaik (AMC), Ies Nijman (Hubrecht), Slavik Koval (EMC) Laurent Francioli (UU), Kai Ye (LUMC), Jeroen Laros (LUMC), Lennart Karssen (EMC), JoukeJan Hottenga (VU), Mathijs Kattenberg (VU), David van Enckvort (NBIC), Leon Mei (NBIC), Elise van Leeuwen (EMC), … and many, many others GoNL Steering group (coordination) Cisca Wijmenga (PI GoNL), Morris Swertz (PI analysis), Gertjan van Ommen (LUMC), Eline Slagboom (LUMC), Jasper Bovenberg (ELSI issues), Cornelia van Duijn (EMC), Dorret Boomsma (VU), Paul de Bakker (co-PI analysis, UU) Get all as open source: GoNL - http://www.nlgenome.nl MOLGENIS - http://www.molgenis.org Analysis team - http://www.bbmriwiki.nl Contact? [email_address]](https://image.slidesharecdn.com/swertzbosc2011-110718162003-phpapp01/75/D02-NextGenSeq-MOLGENIS-50-2048.jpg)