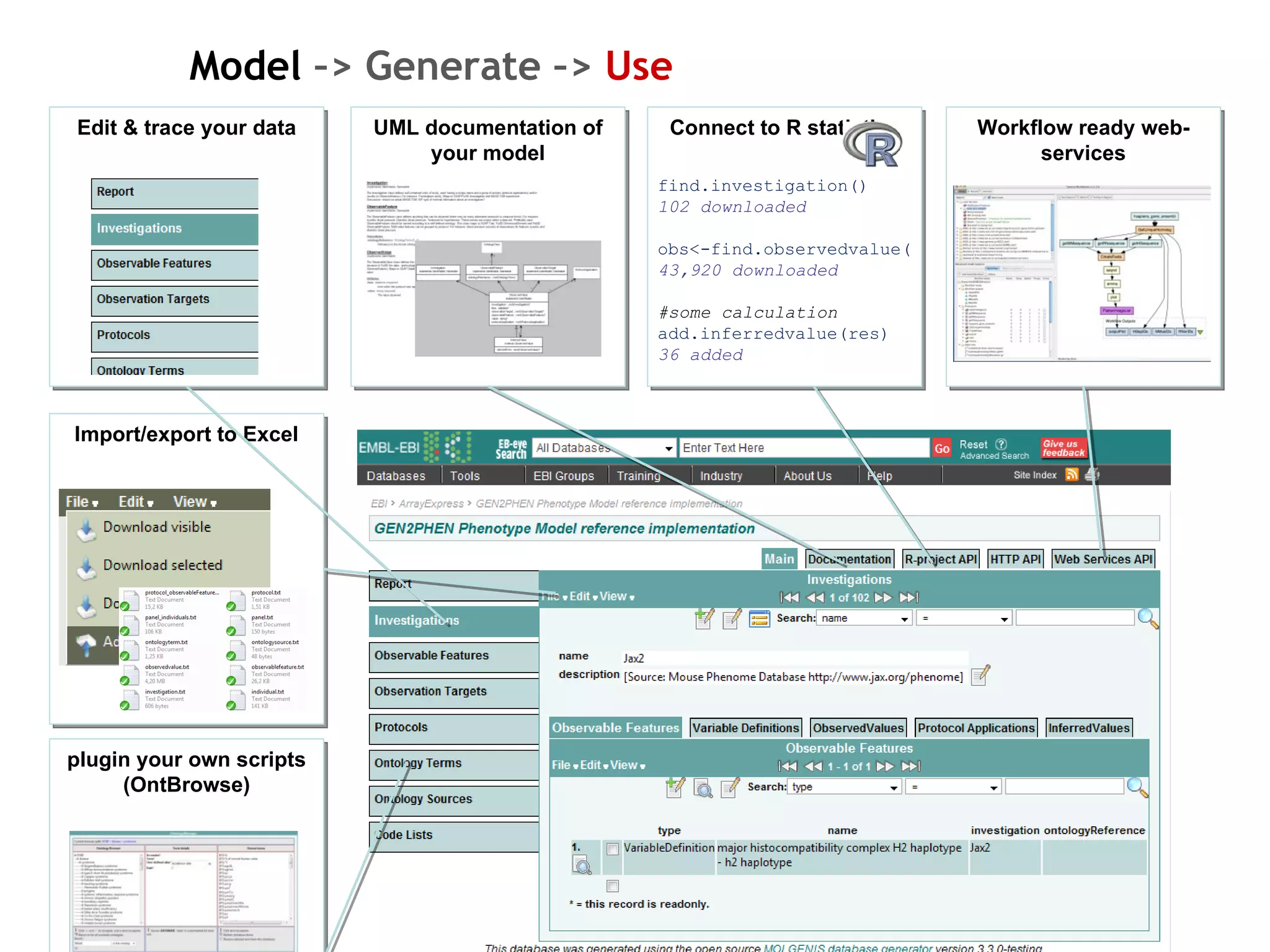
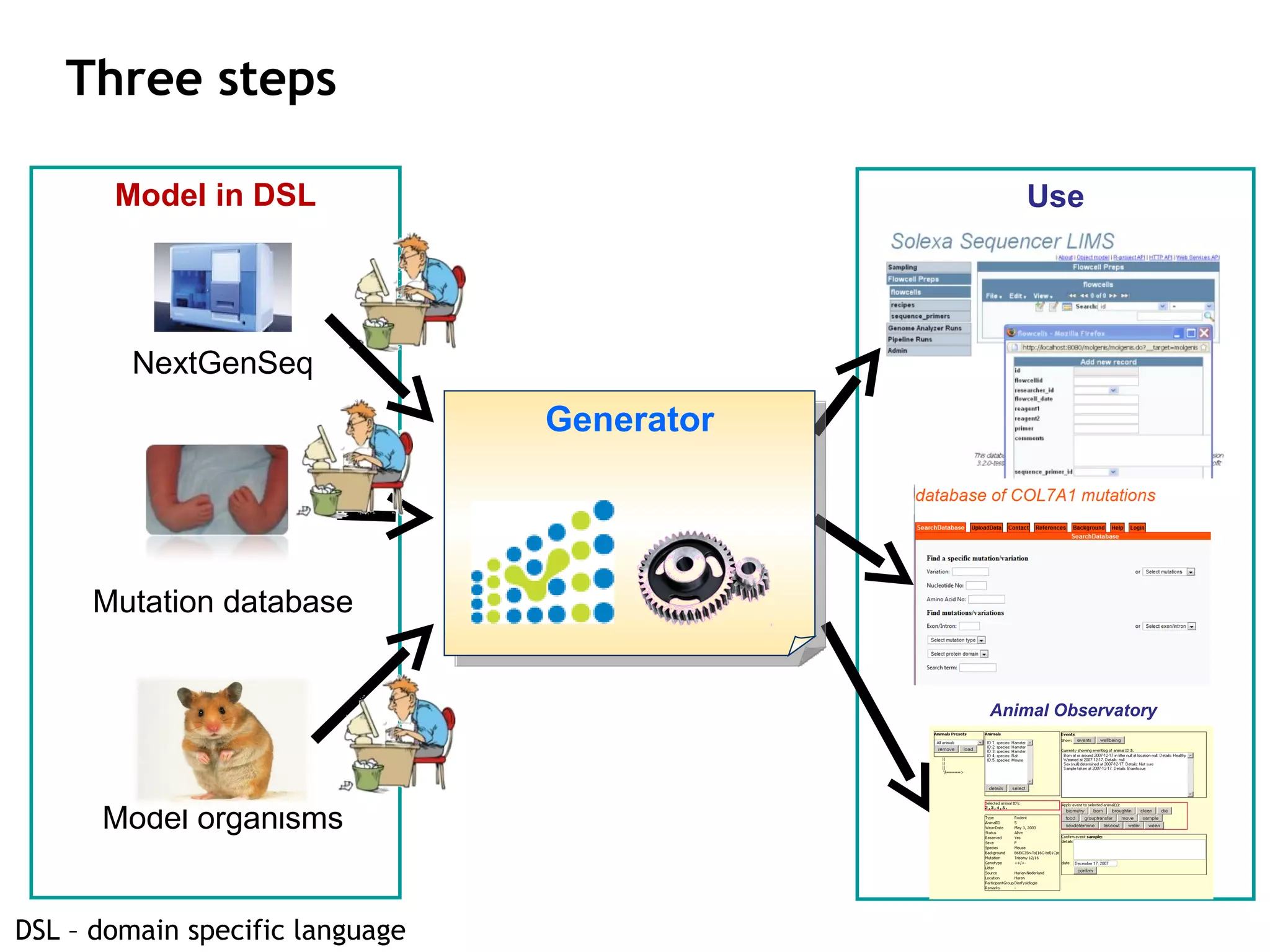
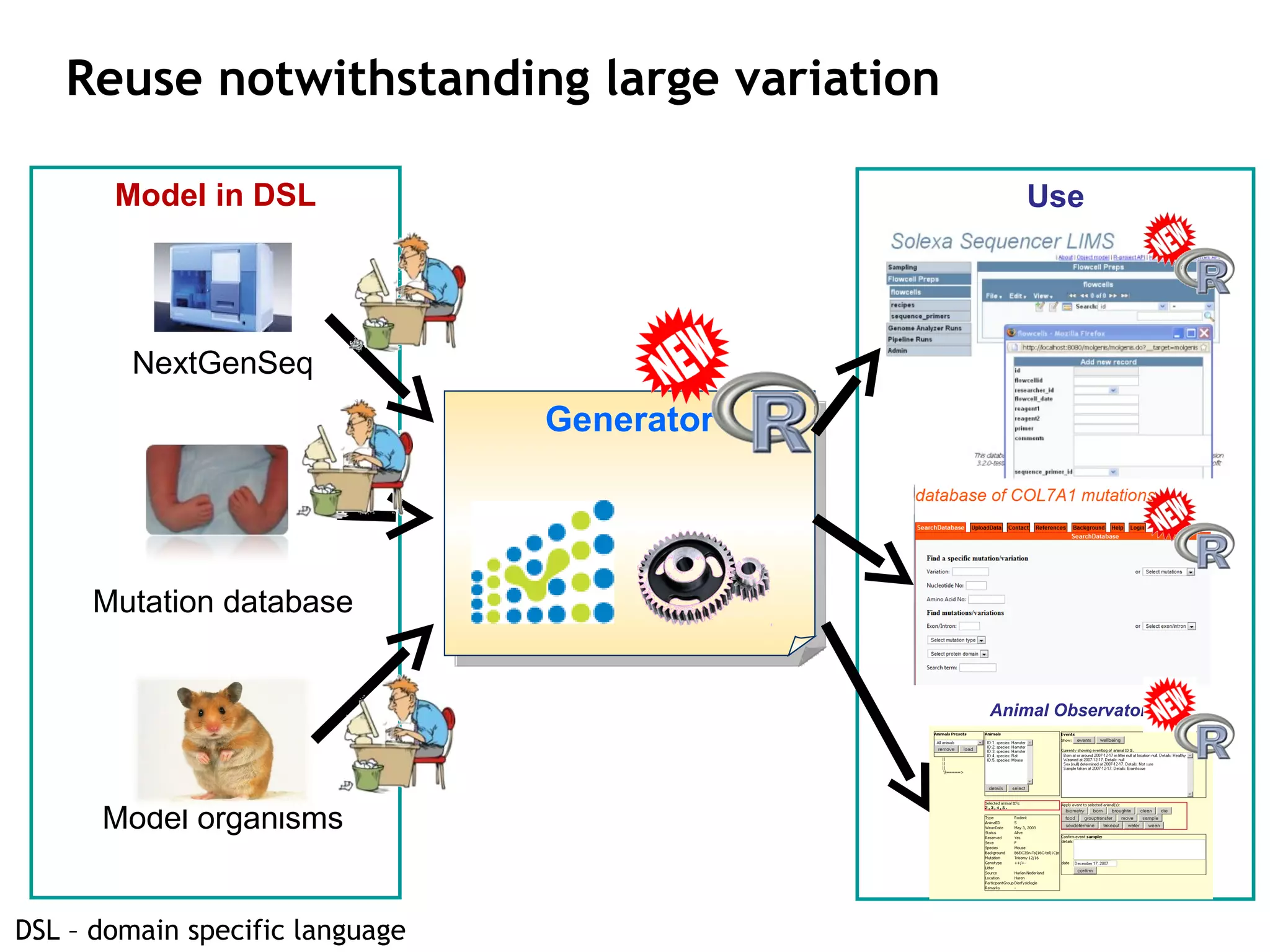
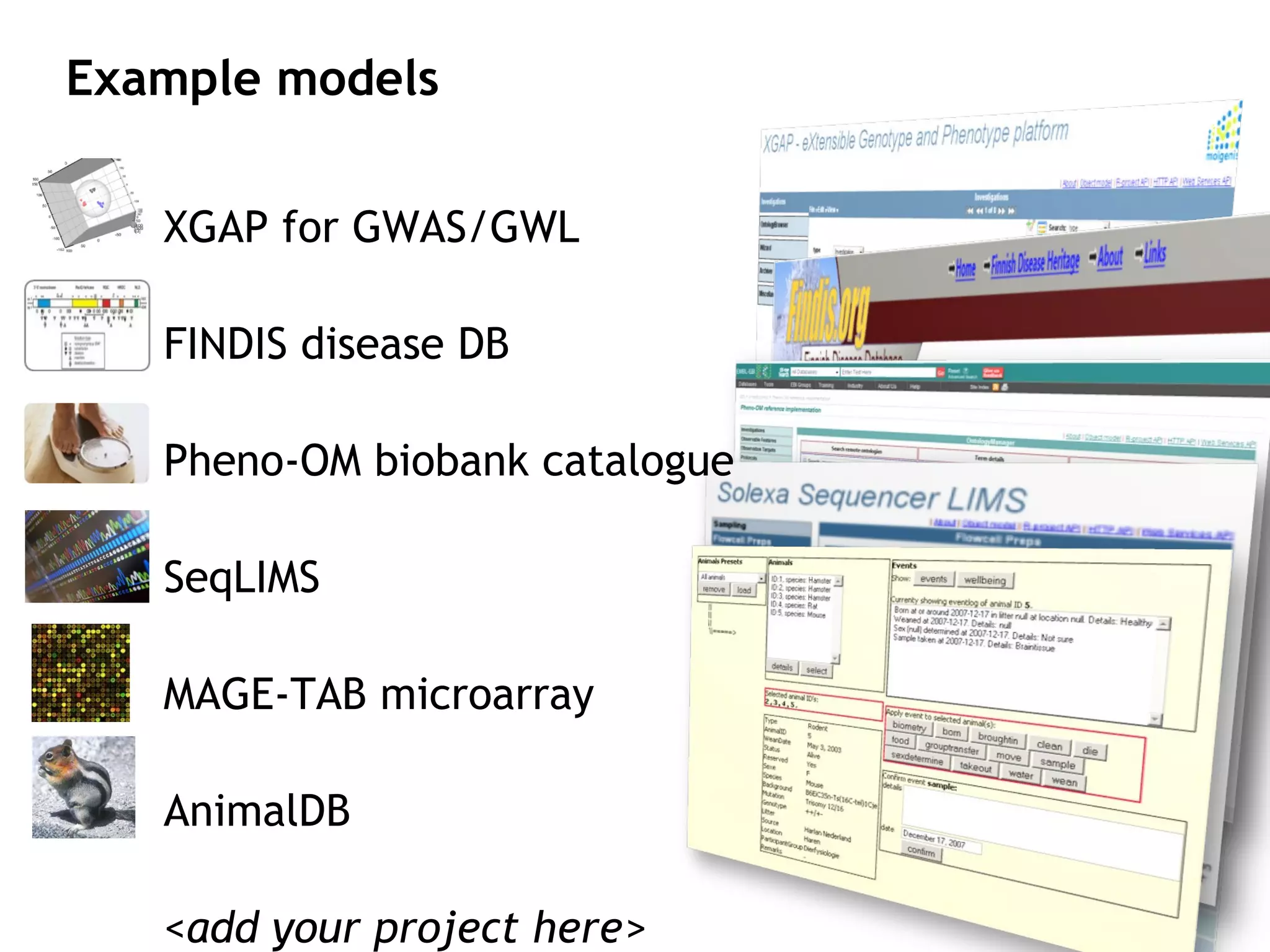
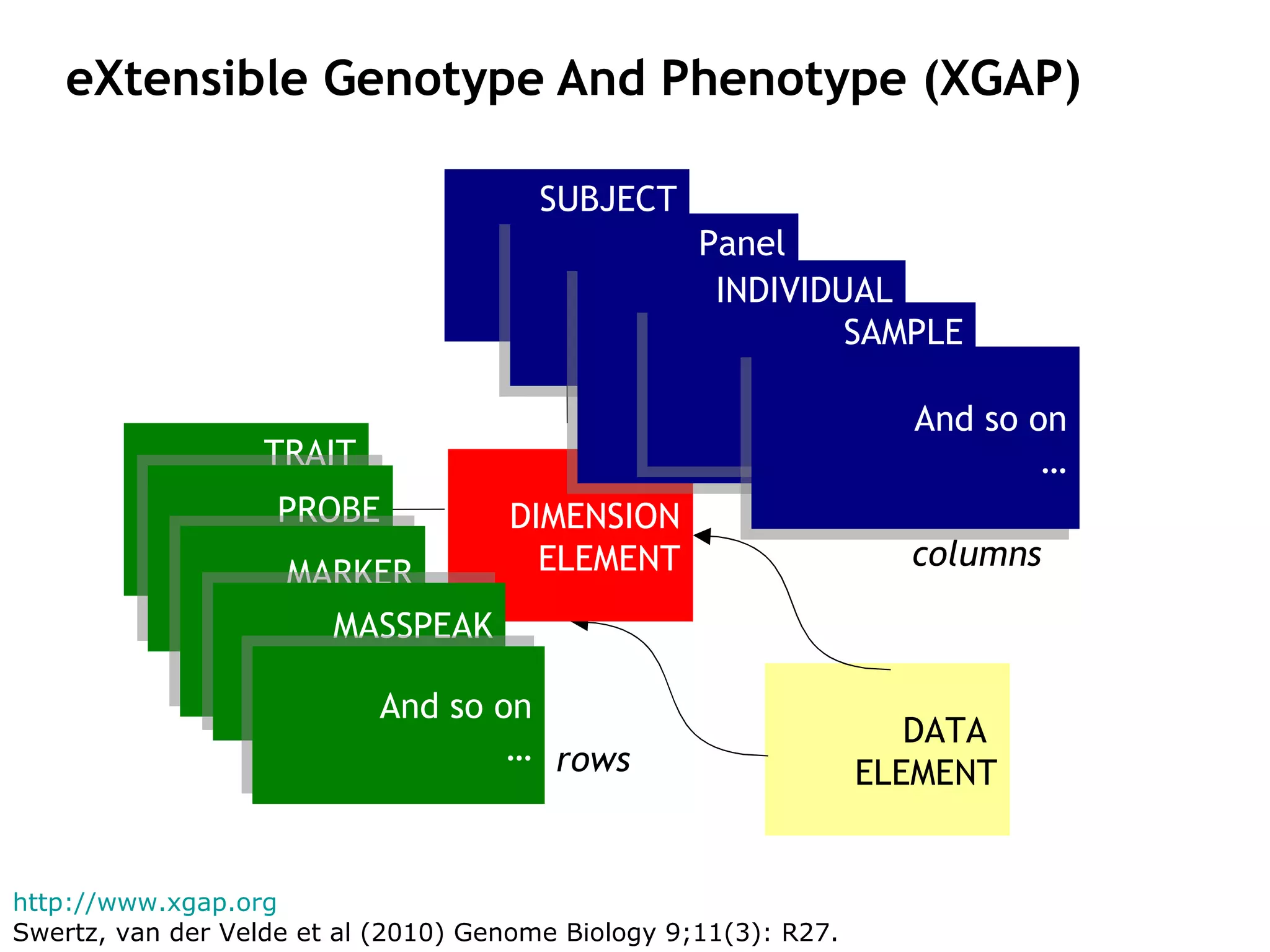
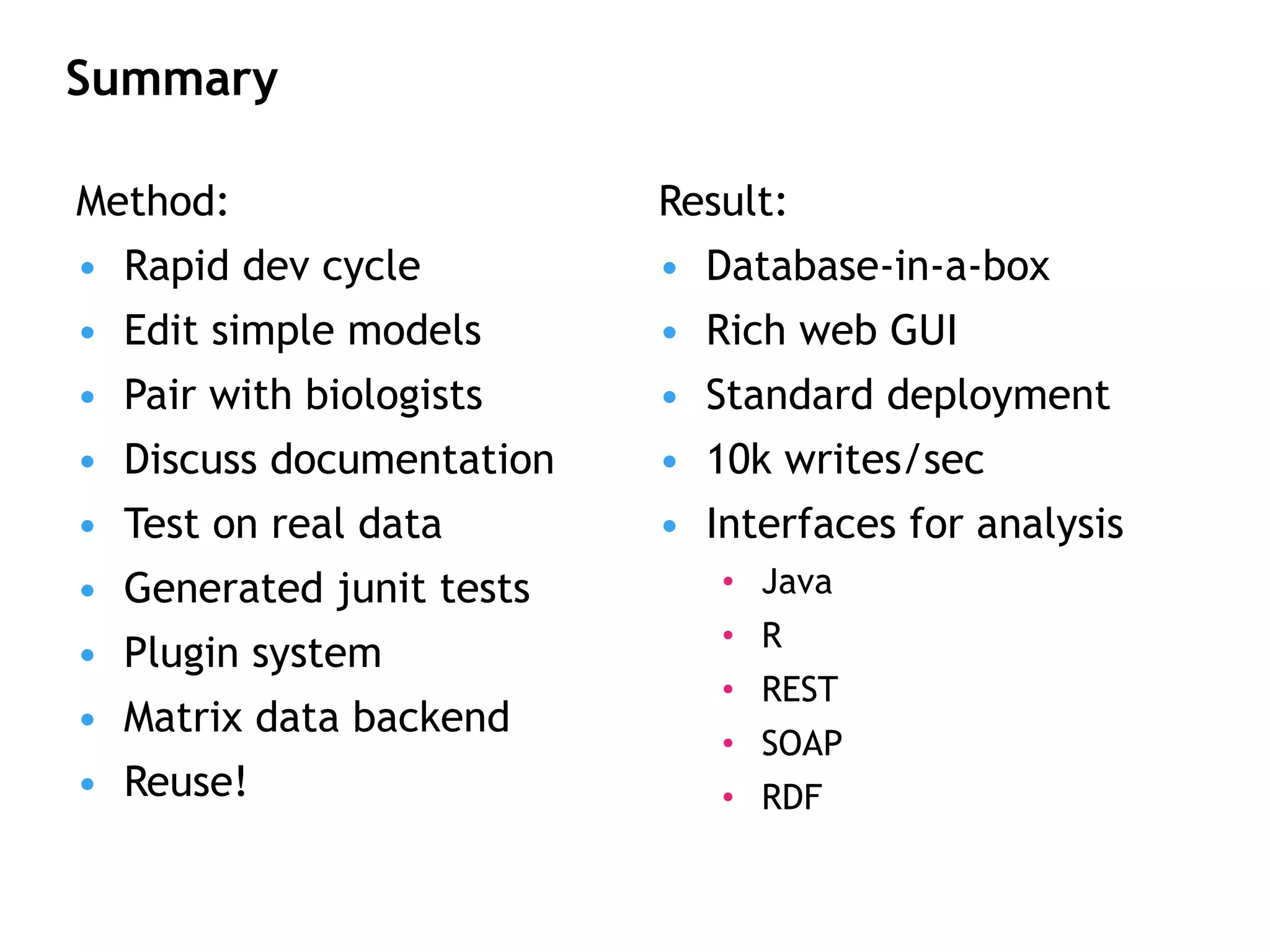
The document describes MOLGENIS, an open-source software system that allows users to define data models and generate full-featured web applications and databases from those models. Key features include a graphical user interface, database integration, support for common data formats, and the ability to rapidly develop applications by editing simple domain-specific models. The system has been applied to build several genomic and biomedical databases.






![ISMB E15 - MOLGENIS Q01 – QTL analysis E19 – MAGE-TAB P05 – OntoCAT TT36, Tuesday 12.15-12.40 Web MOLGENIS: http://www.molgenis.org XGAP: http://www.xgap.org MAGETAB-OM: http://magetab-om.sourceforge.net/ Pheno-OM: http://wwwdev.ebi.ac.uk/microarray-srv/pheno/ FINDIS: http://www.findis.org/molgenis_findis/ AnimalDB: http://www.animaldb.org Pubmed Swertz & Jansen (2007) Nature Reviews Genetics 8, 235-243 Swertz et al (2004) Bioinformatics 20(13), 2075-83 Swertz et al (2010) Genome Biology 9;11(3): R27. Thank you! Questions? [email_address]](https://image.slidesharecdn.com/swertzbosc2010molgenis-100710220424-phpapp02/75/Swertz-bosc2010-molgenis-22-2048.jpg)


![Generates 150 files, 30k lines of Java, SQL and R code + docs 0 INFO [myFactory] working dir: D:\Development\Molgenis33Workspace\molgenis4phenotype 78 INFO [myFactory] MOLGENIS version 3.3.0-testing 94 INFO [myFactory] Using options: model_database = [pheno_db.xml] #File with data structure specification (in MOLGENIS DSL). model_userinterface = pheno_ui.xml #File with user interface specification (in MOLGENIS DSL). Can be same file as model_database output_src = generated/java #Output-directory for the generated project. output_hand = handwritten/java #Output-directory for the generated project. output_sql = generated/sql #Output-directory for the generated sql files. output_doc = WebContent/doc #Output-directory for the generated documentation. output_type = #Output type of the project, either war (for use in tomcat) or jar (standalone). output_web = WebContent #Output-directory for any generated web resources db_driver = com.mysql.jdbc.Driver #Driver of database. Any JDBC compatible driver should work. db_user = molgenis #Username for database. db_password = xxxxxx #Password for database. db_uri = jdbc:mysql://localhost/pheno #Uri of the database. Default: localhost db_filepath = attachedfiles #Path where the database should store file attachements. Default: null db_jndiname = jdbc/molgenisdb #Used to create a JDBC database resource for the application object_relational_mapping = subclass_per_table #Expert option: Choosing OR strategy. Either 'class_per_table', 'subclass_per_table', 'hierarchy_per_table'. Default: class_per_table mapper_implementation = multiquery #Expert option: Choosing wether multiquery is used instead of prepared statements. Default: false exclude_system = true #Expert option: Whether system tables should be excluded from generation. Default: true force_molgenis_package = false #Expert option. Whether the generated package should be 'molgenis' or the name specified in the model. Default: false auth_loginclass = org.molgenis.framework.security.SimpleSecurity #Expert option. verbose = true #This switch turns the verbose-mode on. compile = false #This switch makes the factory also compile (usefull outside IDE). mail_smtp_protocol = #Sets the email protocol, either smtp, smtps or null. Default: null meaning email disabled mail_smtp_hostname = localhost #SMTP host server. Default: localhost mail_smtp_port = 25 #SMTP host server port. Default: 25 mail_smtp_user = #SMTP user for authenticated emailing. Default: null. mail_smtp_password = #SMTP user for authenticated emailing. Default: null. 110 INFO [MolgenisLanguage] parsing db-schema from [pheno_db.xml] 780 WARN [Entity] [WARNING]: missing key 0 for entity Nameable 780 WARN [Entity] [WARNING]: missing key 0 for entity Nameable 780 WARN [Entity] [WARNING]: missing key 0 for entity Nameable 780 WARN [Entity] [WARNING]: missing key 0 for entity Nameable 780 WARN [Entity] [WARNING]: missing key 0 for entity Nameable 797 WARN [Entity] [WARNING]: missing key 0 for entity Nameable 797 WARN [Entity] [WARNING]: missing key 0 for entity Nameable 844 INFO [MolgenisLanguage] parsing ui-schema 937 INFO [main] generating .... 1717 INFO [TableDocGen] generated WebContent\doc\tabledoc.html 2076 INFO [EntityDocGen] generated WebContent\doc\objectmodel.html 2436 INFO [DotDocGen] generated WebContent\doc\entity-uml-diagram.dot 2545 INFO [DotDocGen] generated WebContent\doc\entity-uml-diagram-pheno.system.dot 2748 INFO [DotDocGen] generated WebContent\doc\entity-uml-diagram-pheno.observation.dot 2842 INFO [DotDocGen] generated WebContent\doc\entity-uml-diagram-pheno.target.dot 2998 INFO [DotDocGen] generated WebContent\doc\entity-uml-diagram-pheno.variable.dot 3138 INFO [DotDocGen] generated WebContent\doc\entity-uml-diagram-pheno.protocol.dot 3997 INFO [DotDocMinimalGen] generated WebContent\doc\entity-uml-minimal-diagram.dot 4184 INFO [DotDocMinimalGen] generated WebContent\doc\entity-uml-diagram-minimal-pheno.system.dot 4388 INFO [DotDocMinimalGen] generated WebContent\doc\entity-uml-diagram-minimal-pheno.observation.dot 4606 INFO [DotDocMinimalGen] generated WebContent\doc\entity-uml-diagram-minimal-pheno.target.dot 4731 INFO [DotDocMinimalGen] generated WebContent\doc\entity-uml-diagram-minimal-pheno.variable.dot 4887 INFO [DotDocMinimalGen] generated WebContent\doc\entity-uml-diagram-minimal-pheno.protocol.dot 5184 INFO [ClassDocGen] generated WebContent\doc\classmodel.html 5293 INFO [InMemoryDatabaseGen] generated generated\java\ui\data\InMemoryDatabase.java 5609 INFO [MySqlCreateSubclassPerTableGen] generated generated\sql\create_tables.sql 5671 INFO [JDBCDatabaseGen] generated generated\java\ui\JDBCDatabase.java 5921 INFO [DataTypeGen] generated generated\java\pheno\core\data\types\Identifiable.java 5921 INFO [DataTypeGen] generated generated\java\pheno\core\data\types\Nameable.java 5968 INFO [DataTypeGen] generated generated\java\pheno\core\data\types\OntologySource.java 6014 INFO [DataTypeGen] generated generated\java\pheno\core\data\types\OntologyTerm.java 6030 INFO [DataTypeGen] generated generated\java\pheno\core\data\types\Investigation.java 6061 INFO [DataTypeGen] generated generated\java\pheno\core\data\types\ObservableFeature.java 6124 INFO [DataTypeGen] generated generated\java\pheno\core\data\types\ObservedValue.java 6170 INFO [DataTypeGen] generated generated\java\pheno\core\data\types\ObservedRelationship.java 6217 INFO [DataTypeGen] generated generated\java\pheno\core\data\types\InferredValue.java 6233 INFO [DataTypeGen] generated generated\java\pheno\core\data\types\ObservationTarget.java 6280 INFO [DataTypeGen] generated generated\java\pheno\core\data\types\Individual.java 6311 INFO [DataTypeGen] generated generated\java\pheno\core\data\types\Panel.java 6326 INFO [DataTypeGen] generated generated\java\pheno\core\data\types\CodeList.java 6327 INFO [DataTypeGen] generated generated\java\pheno\core\data\types\Code.java 6374 INFO [DataTypeGen] generated generated\java\pheno\core\data\types\Protocol.java 6390 INFO [DataTypeGen] generated generated\java\pheno\core\data\types\ProtocolApplication.java 6405 INFO [DataTypeGen] generated generated\java\pheno\core\data\types\ProtocolParameter.java 6437 INFO [DataTypeGen] generated generated\java\pheno\core\data\types\ParameterValue.java 6452 INFO [DataTypeGen] generated generated\java\pheno\core\data\types\InferredValue_derivedFrom.java 6468 INFO [DataTypeGen] generated generated\java\pheno\core\data\types\Panel_individuals.java 6483 INFO [DataTypeGen] generated generated\java\pheno\core\data\types\Protocol_observableFeatures.java 6499 INFO [DataTypeGen] generated generated\java\pheno\core\data\types\Protocol_protocolComponents.java 6624 INFO [MultiqueryMapperGen] generated generated\java\pheno\core\data\mappers\OntologySourceMapper.java 6655 INFO [MultiqueryMapperGen] generated generated\java\pheno\core\data\mappers\OntologyTermMapper.java 6671 INFO [MultiqueryMapperGen] generated generated\java\pheno\core\data\mappers\InvestigationMapper.java 6702 INFO [MultiqueryMapperGen] generated generated\java\pheno\core\data\mappers\ObservableFeatureMapper.java 6733 INFO [MultiqueryMapperGen] generated generated\java\pheno\core\data\mappers\ObservedValueMapper.java 6780 INFO [MultiqueryMapperGen] generated generated\java\pheno\core\data\mappers\ObservedRelationshipMapper.java 6827 INFO [MultiqueryMapperGen] generated generated\java\pheno\core\data\mappers\InferredValueMapper.java 6842 INFO [MultiqueryMapperGen] generated generated\java\pheno\core\data\mappers\ObservationTargetMapper.java 6873 INFO [MultiqueryMapperGen] generated generated\java\pheno\core\data\mappers\IndividualMapper.java 6889 INFO [MultiqueryMapperGen] generated generated\java\pheno\core\data\mappers\PanelMapper.java 6905 INFO [MultiqueryMapperGen] generated generated\java\pheno\core\data\mappers\CodeListMapper.java 6936 INFO [MultiqueryMapperGen] generated generated\java\pheno\core\data\mappers\CodeMapper.java 6951 INFO [MultiqueryMapperGen] generated generated\java\pheno\core\data\mappers\ProtocolMapper.java 6983 INFO [MultiqueryMapperGen] generated generated\java\pheno\core\data\mappers\ProtocolApplicationMapper.java 6998 INFO [MultiqueryMapperGen] generated generated\java\pheno\core\data\mappers\ProtocolParameterMapper.java 7029 INFO [MultiqueryMapperGen] generated generated\java\pheno\core\data\mappers\ParameterValueMapper.java 7045 INFO [MultiqueryMapperGen] generated generated\java\pheno\core\data\mappers\InferredValue_derivedFromMapper.java 7061 INFO [MultiqueryMapperGen] generated generated\java\pheno\core\data\mappers\Panel_individualsMapper.java 7076 INFO [MultiqueryMapperGen] generated generated\java\pheno\core\data\mappers\Protocol_observableFeaturesMapper.java 7092 INFO [MultiqueryMapperGen] generated generated\java\pheno\core\data\mappers\Protocol_protocolComponentsMapper.java 7217 INFO [JDBCMetaDatabaseGen] generated generated\java\ui\JDBCMetaDatabase.java 7263 INFO [CountPerEntityGen] generated generated\sql\count_per_entity.sql 7310 INFO [CountPerTableGen] generated generated\sql\count_per_table.sql 7341 INFO [FillMetadataTablesGen] generated generated\sql\insert_metadata.sql 7405 INFO [CsvReaderGen] generated generated\java\pheno\core\data\csv\OntologySourceCsvReader.java 7420 INFO [CsvReaderGen] generated generated\java\pheno\core\data\csv\OntologyTermCsvReader.java 7420 INFO [CsvReaderGen] generated generated\java\pheno\core\data\csv\InvestigationCsvReader.java 7436 INFO [CsvReaderGen] generated generated\java\pheno\core\data\csv\ObservableFeatureCsvReader.java 7452 INFO [CsvReaderGen] generated generated\java\pheno\core\data\csv\ObservedValueCsvReader.java 7467 INFO [CsvReaderGen] generated generated\java\pheno\core\data\csv\ObservedRelationshipCsvReader.java 7483 INFO [CsvReaderGen] generated generated\java\pheno\core\data\csv\InferredValueCsvReader.java 7498 INFO [CsvReaderGen] generated generated\java\pheno\core\data\csv\ObservationTargetCsvReader.java 7514 INFO [CsvReaderGen] generated generated\java\pheno\core\data\csv\IndividualCsvReader.java 7514 INFO [CsvReaderGen] generated generated\java\pheno\core\data\csv\PanelCsvReader.java 7530 INFO [CsvReaderGen] generated generated\java\pheno\core\data\csv\CodeListCsvReader.java 7545 INFO [CsvReaderGen] generated generated\java\pheno\core\data\csv\CodeCsvReader.java 7545 INFO [CsvReaderGen] generated generated\java\pheno\core\data\csv\ProtocolCsvReader.java 7561 INFO [CsvReaderGen] generated generated\java\pheno\core\data\csv\ProtocolApplicationCsvReader.java 7561 INFO [CsvReaderGen] generated generated\java\pheno\core\data\csv\ProtocolParameterCsvReader.java 7576 INFO [CsvReaderGen] generated generated\java\pheno\core\data\csv\ParameterValueCsvReader.java 7576 INFO [CsvReaderGen] generated generated\java\pheno\core\data\csv\InferredValue_derivedFromCsvReader.java 7592 INFO [CsvReaderGen] generated generated\java\pheno\core\data\csv\Panel_individualsCsvReader.java 7608 INFO [CsvReaderGen] generated generated\java\pheno\core\data\csv\Protocol_observableFeaturesCsvReader.java 7608 INFO [CsvReaderGen] generated generated\java\pheno\core\data\csv\Protocol_protocolComponentsCsvReader.java 7748 INFO [REntityGen] generated generated\java\pheno\core\R\OntologySource.R 7748 INFO [REntityGen] generated generated\java\pheno\core\R\OntologyTerm.R 7764 INFO [REntityGen] generated generated\java\pheno\core\R\Investigation.R 7779 INFO [REntityGen] generated generated\java\pheno\core\R\ObservableFeature.R 7779 INFO [REntityGen] generated generated\java\pheno\core\R\ObservedValue.R 7795 INFO [REntityGen] generated generated\java\pheno\core\R\ObservedRelationship.R 7795 INFO [REntityGen] generated generated\java\pheno\core\R\InferredValue.R 7810 INFO [REntityGen] generated generated\java\pheno\core\R\ObservationTarget.R 7810 INFO [REntityGen] generated generated\java\pheno\core\R\Individual.R 7826 INFO [REntityGen] generated generated\java\pheno\core\R\Panel.R 7826 INFO [REntityGen] generated generated\java\pheno\core\R\CodeList.R 7842 INFO [REntityGen] generated generated\java\pheno\core\R\Code.R 7857 INFO [REntityGen] generated generated\java\pheno\core\R\Protocol.R 7857 INFO [REntityGen] generated generated\java\pheno\core\R\ProtocolApplication.R 7873 INFO [REntityGen] generated generated\java\pheno\core\R\ProtocolParameter.R 7873 INFO [REntityGen] generated generated\java\pheno\core\R\ParameterValue.R 7888 INFO [REntityGen] generated generated\java\pheno\core\R\InferredValue_derivedFrom.R 7888 INFO [REntityGen] generated generated\java\pheno\core\R\Panel_individuals.R 7888 INFO [REntityGen] generated generated\java\pheno\core\R\Protocol_observableFeatures.R 7904 INFO [REntityGen] generated generated\java\pheno\core\R\Protocol_protocolComponents.R 7998 INFO [RApi] generated generated\java\source.R 8044 INFO [HtmlFormGen] generated generated\java\pheno\core\html\IdentifiableHtmlForm.java 8044 INFO [HtmlFormGen] generated generated\java\pheno\core\html\NameableHtmlForm.java 8044 INFO [HtmlFormGen] generated generated\java\pheno\core\html\OntologySourceHtmlForm.java 8044 INFO [HtmlFormGen] generated generated\java\pheno\core\html\OntologyTermHtmlForm.java 8060 INFO [HtmlFormGen] generated generated\java\pheno\core\html\InvestigationHtmlForm.java 8060 INFO [HtmlFormGen] generated generated\java\pheno\core\html\ObservableFeatureHtmlForm.java 8076 INFO [HtmlFormGen] generated generated\java\pheno\core\html\ObservedValueHtmlForm.java 8076 INFO [HtmlFormGen] generated generated\java\pheno\core\html\ObservedRelationshipHtmlForm.java 8076 INFO [HtmlFormGen] generated generated\java\pheno\core\html\InferredValueHtmlForm.java 8091 INFO [HtmlFormGen] generated generated\java\pheno\core\html\ObservationTargetHtmlForm.java 8091 INFO [HtmlFormGen] generated generated\java\pheno\core\html\IndividualHtmlForm.java 8091 INFO [HtmlFormGen] generated generated\java\pheno\core\html\PanelHtmlForm.java 8091 INFO [HtmlFormGen] generated generated\java\pheno\core\html\CodeListHtmlForm.java 8107 INFO [HtmlFormGen] generated generated\java\pheno\core\html\CodeHtmlForm.java 8107 INFO [HtmlFormGen] generated generated\java\pheno\core\html\ProtocolHtmlForm.java 8107 INFO [HtmlFormGen] generated generated\java\pheno\core\html\ProtocolApplicationHtmlForm.java 8107 INFO [HtmlFormGen] generated generated\java\pheno\core\html\ProtocolParameterHtmlForm.java 8122 INFO [HtmlFormGen] generated generated\java\pheno\core\html\ParameterValueHtmlForm.java 8122 INFO [HtmlFormGen] generated generated\java\pheno\core\html\InferredValue_derivedFromHtmlForm.java 8122 INFO [HtmlFormGen] generated generated\java\pheno\core\html\Panel_individualsHtmlForm.java 8122 INFO [HtmlFormGen] generated generated\java\pheno\core\html\Protocol_observableFeaturesHtmlForm.java 8138 INFO [HtmlFormGen] generated generated\java\pheno\core\html\Protocol_protocolComponentsHtmlForm.java 8138 INFO [MolgenisServletContextGen] generated WebContent\META-INF\context.xml 8169 INFO [MolgenisContextListenerGen] generated generated\java\servlet\ContextListener.java 8232 INFO [MolgenisServletGen] generated generated\java\MolgenisServlet.java 8403 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\InvestigationsForm.java 8560 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\Investigations\InvestigationMenu\ObservableFeaturesForm.java 8591 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\Investigations\InvestigationMenu\PanelsForm.java 8654 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\Investigations\InvestigationMenu\Panels\IndividualsForm.java 8701 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\Investigations\InvestigationMenu\ObservedValuesForm.java 8732 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\Investigations\InvestigationMenu\ProtocolApplicationsForm.java 8825 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\Investigations\InvestigationMenu\ProtocolApplications\ProtocolApplicationMenu\ParameterValuesForm.java 8857 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\Investigations\InvestigationMenu\ProtocolApplications\ProtocolApplicationMenu\ObservedValuesForm.java 8888 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\Investigations\InvestigationMenu\ProtocolApplications\ProtocolApplicationMenu\InferredValuesForm.java 9013 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\Investigations\InvestigationMenu\InferredValuesForm.java 9044 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\ObservableFeaturesForm.java 9137 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\ObservationTargets\IndividualsForm.java 9169 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\ObservationTargets\PanelsForm.java 9200 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\ProtocolsForm.java 9293 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\Protocols\ProtocolMenu\ParametersForm.java 9325 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\Protocols\ProtocolMenu\ProtocolComponentsForm.java 9496 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\Ontologies\OntologyTermsForm.java 9528 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\Ontologies\OntologySourcesForm.java 9606 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\Ontologies\OntologySources\OntologyTermsForm.java 9638 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\Ontologies\CodeListsForm.java 9700 INFO [FormScreenGen] generated generated\java\ui\screen\TopMenu\Main\Ontologies\CodeLists\CodesForm.java 9965 INFO [MenuScreenGen] generated generated\java\ui\screen\TopMenuMenu.java 10012 INFO [MenuScreenGen] generated generated\java\ui\screen\TopMenu\MainMenu.java 10059 INFO [MenuScreenGen] generated generated\java\ui\screen\TopMenu\Main\Investigations\InvestigationMenuMenu.java 10152 INFO [MenuScreenGen] generated generated\java\ui\screen\TopMenu\Main\Investigations\InvestigationMenu\ProtocolApplications\ProtocolApplicationMenuMenu.java 10230 INFO [MenuScreenGen] generated generated\java\ui\screen\TopMenu\Main\ObservationTargetsMenu.java 10293 INFO [MenuScreenGen] generated generated\java\ui\screen\TopMenu\Main\Protocols\ProtocolMenuMenu.java 10324 INFO [MenuScreenGen] generated generated\java\ui\screen\TopMenu\Main\OntologiesMenu.java 11354 INFO [PluginScreenGen] generated Molgenis33Workspace\molgenis4phenotype\generated\java\ui\screen\TopMenu\Main\ReportPlugin.java 11557 INFO [PluginScreenGen] generated Molgenis33Workspace\molgenis4phenotype\generated\java\ui\screen\TopMenu\Main\Ontologies\OntologyManagerPlugin.java 11604 INFO [PluginScreenGen] generated Molgenis33Workspace\molgenis4phenotype\generated\java\ui\screen\TopMenu\Model_documentationPlugin.java 11604 INFO [PluginScreenGen] generated Molgenis33Workspace\molgenis4phenotype\generated\java\ui\screen\TopMenu\RprojectApiPlugin.java 11620 INFO [PluginScreenGen] generated Molgenis33Workspace\molgenis4phenotype\generated\java\ui\screen\TopMenu\HttpApiPlugin.java 11635 INFO [PluginScreenGen] generated Molgenis33Workspace\molgenis4phenotype\generated\java\ui\screen\TopMenu\WebServicesApiPlugin.java 11651 WARN [PluginScreenFTLTemplateGen] Skipped because exists: handwritten\java\plugin\report\InvestigationOverview.ftl 11807 WARN [PluginScreenFTLTemplateGen] Skipped because exists: handwritten\java\plugin\OntologyBrowser\OntologyBrowserPlugin.ftl 11807 WARN [PluginScreenFTLTemplateGen] Skipped because exists: handwritten\java\plugin\topmenu\DocumentationScreen.ftl 11807 WARN [PluginScreenFTLTemplateGen] Skipped because exists: handwritten\java\plugin\topmenu\RprojectApiScreen.ftl 11823 WARN [PluginScreenFTLTemplateGen] Skipped because exists: handwritten\java\plugin\topmenu\HttpAPiScreen.ftl 11823 WARN [PluginScreenFTLTemplateGen] Skipped because exists: handwritten\java\plugin\topmenu\SoapApiScreen.ftl 11854 WARN [PluginScreenJavaTemplateGen] Skipped because exists: handwritten\java\plugin\report\InvestigationOverview.java 12057 WARN [PluginScreenJavaTemplateGen] Skipped because exists: handwritten\java\plugin\OntologyBrowser\OntologyBrowserPlugin.java 12072 WARN [PluginScreenJavaTemplateGen] Skipped because exists: handwritten\java\plugin\topmenu\DocumentationScreen.java 12088 WARN [PluginScreenJavaTemplateGen] Skipped because exists: handwritten\java\plugin\topmenu\RprojectApiScreen.java 12088 WARN [PluginScreenJavaTemplateGen] Skipped because exists: handwritten\java\plugin\topmenu\HttpAPiScreen.java 12088 WARN [PluginScreenJavaTemplateGen] Skipped because exists: handwritten\java\plugin\topmenu\SoapApiScreen.java 12103 INFO [MolgenisServletContextGen] generated WebContent\META-INF\context.xml 12259 INFO [SoapApiGen] generated generated\java\ui\SoapApi.java 12353 INFO [CsvExportGen] generated generated\java\tools\CsvExport.java 12431 INFO [CsvImportByNameGen] generated generated\java\tools\CsvImportByName.java 12636 INFO [CopyMemoryToDatabaseGen] generated generated\java\ui\tools\CopyMemoryToDatabase.java Model –> Generate –> Use](https://image.slidesharecdn.com/swertzbosc2010molgenis-100710220424-phpapp02/75/Swertz-bosc2010-molgenis-25-2048.jpg)