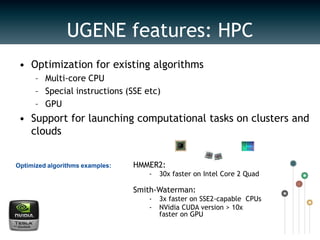
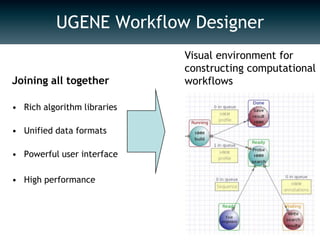
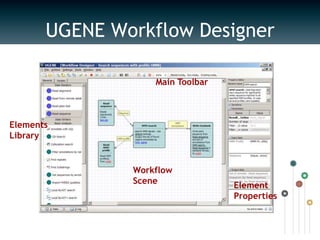
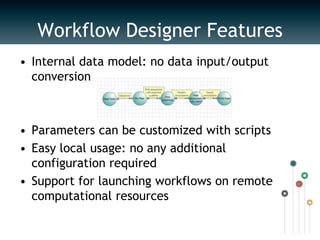
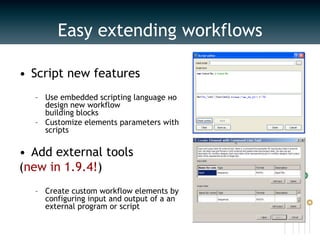
UGENE is a multiplatform open-source toolkit for complex genome analysis. It integrates popular bioinformatics tools into a unified visual and computational solution. Written in C++/Qt, UGENE has a modular structure and integrated plugin system. It supports over 20 common biological data formats and allows users to retrieve information from remote databases. UGENE features rich visualization capabilities and algorithms optimized for multi-core CPUs and GPUs. The UGENE Workflow Designer provides a visual environment for constructing computational workflows combining algorithms, data formats, and high performance capabilities.