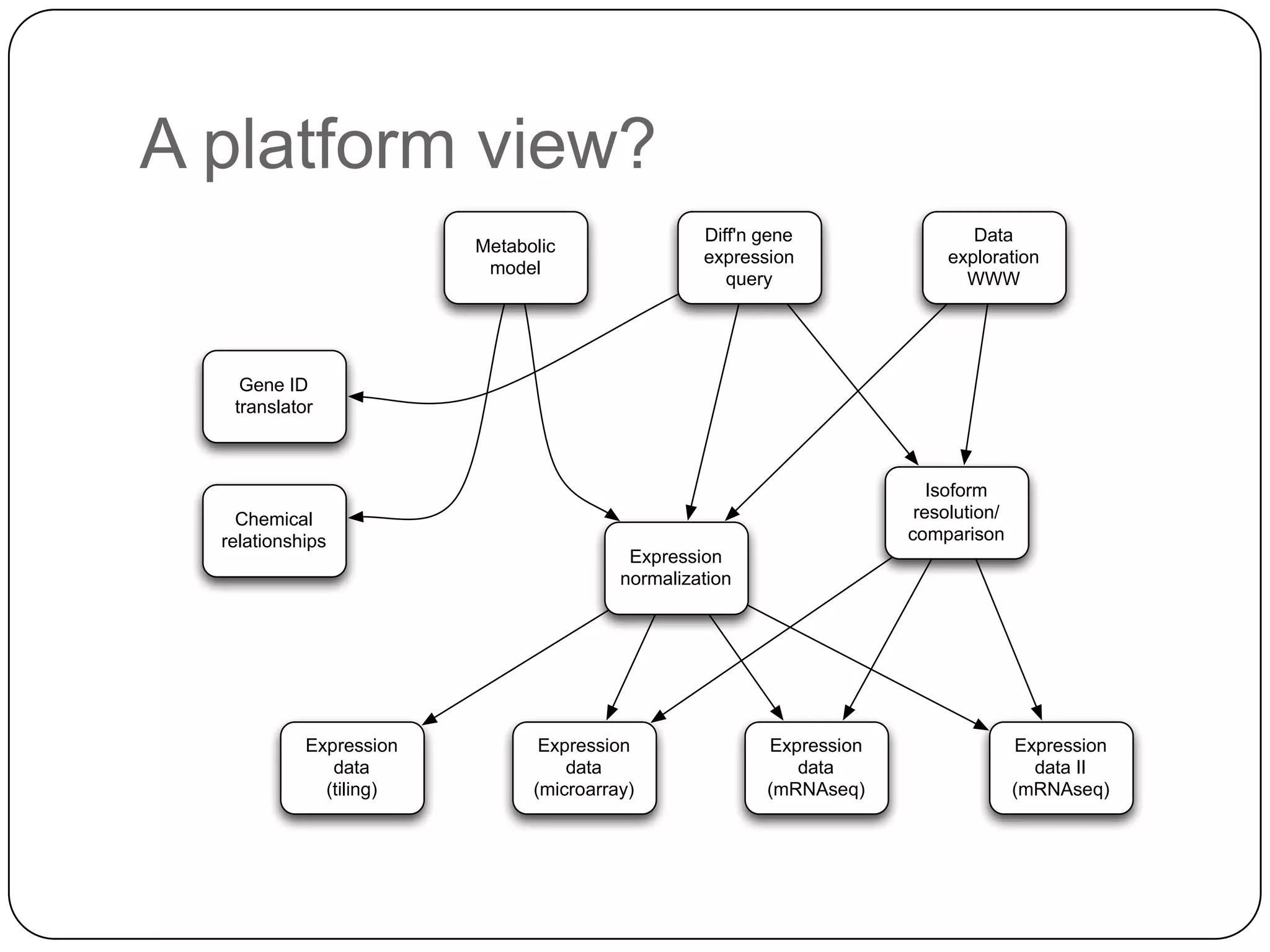
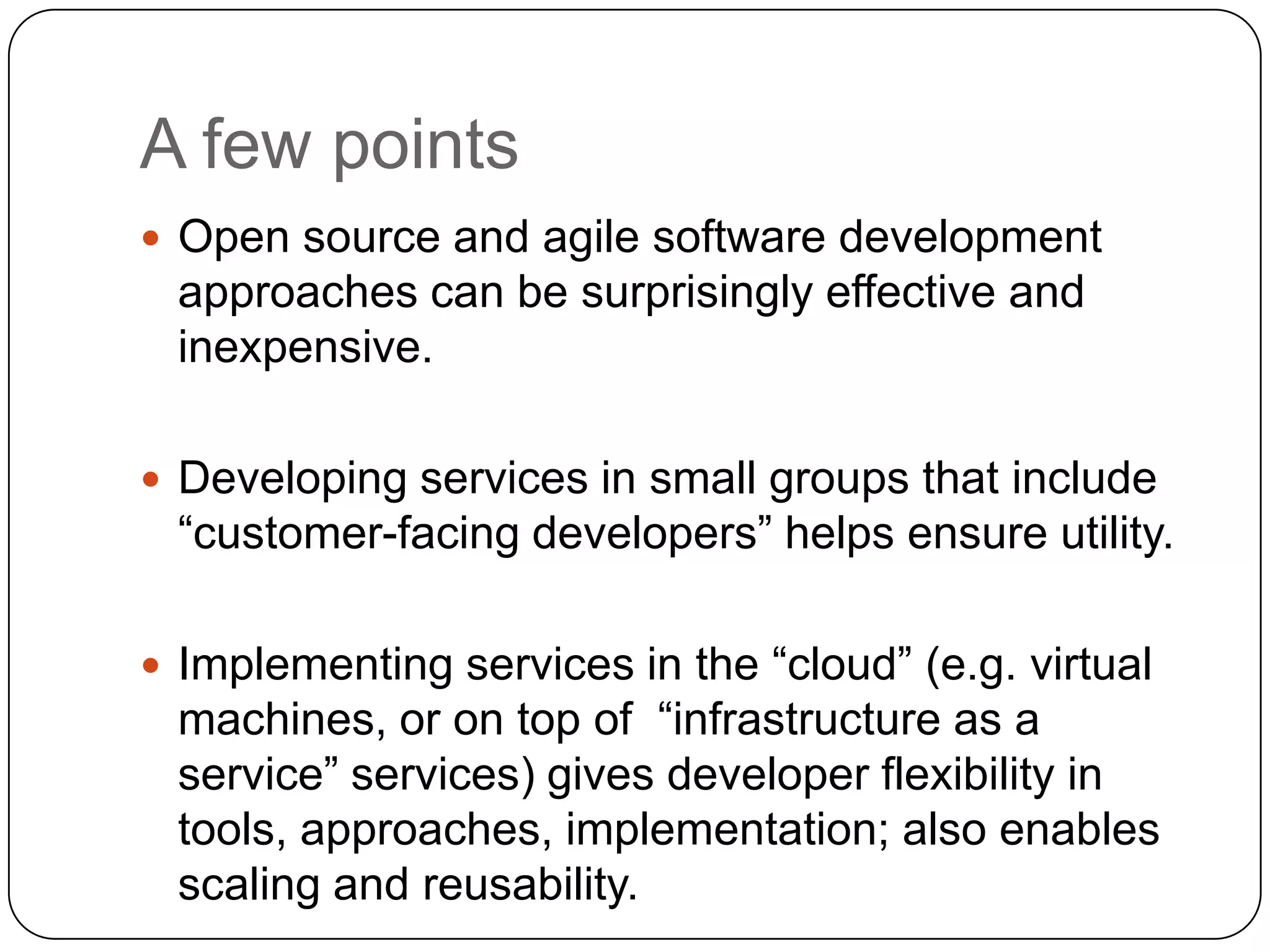
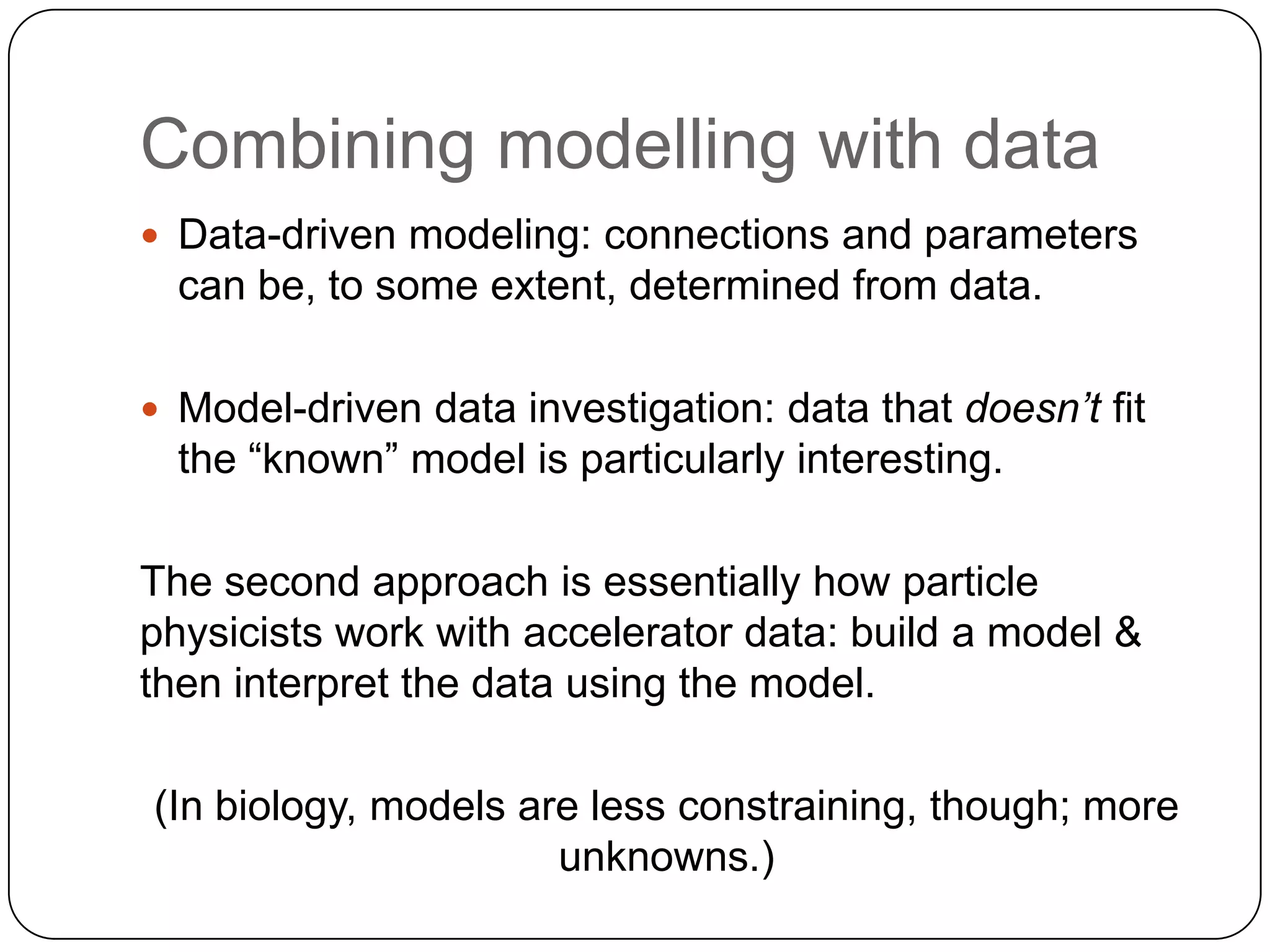
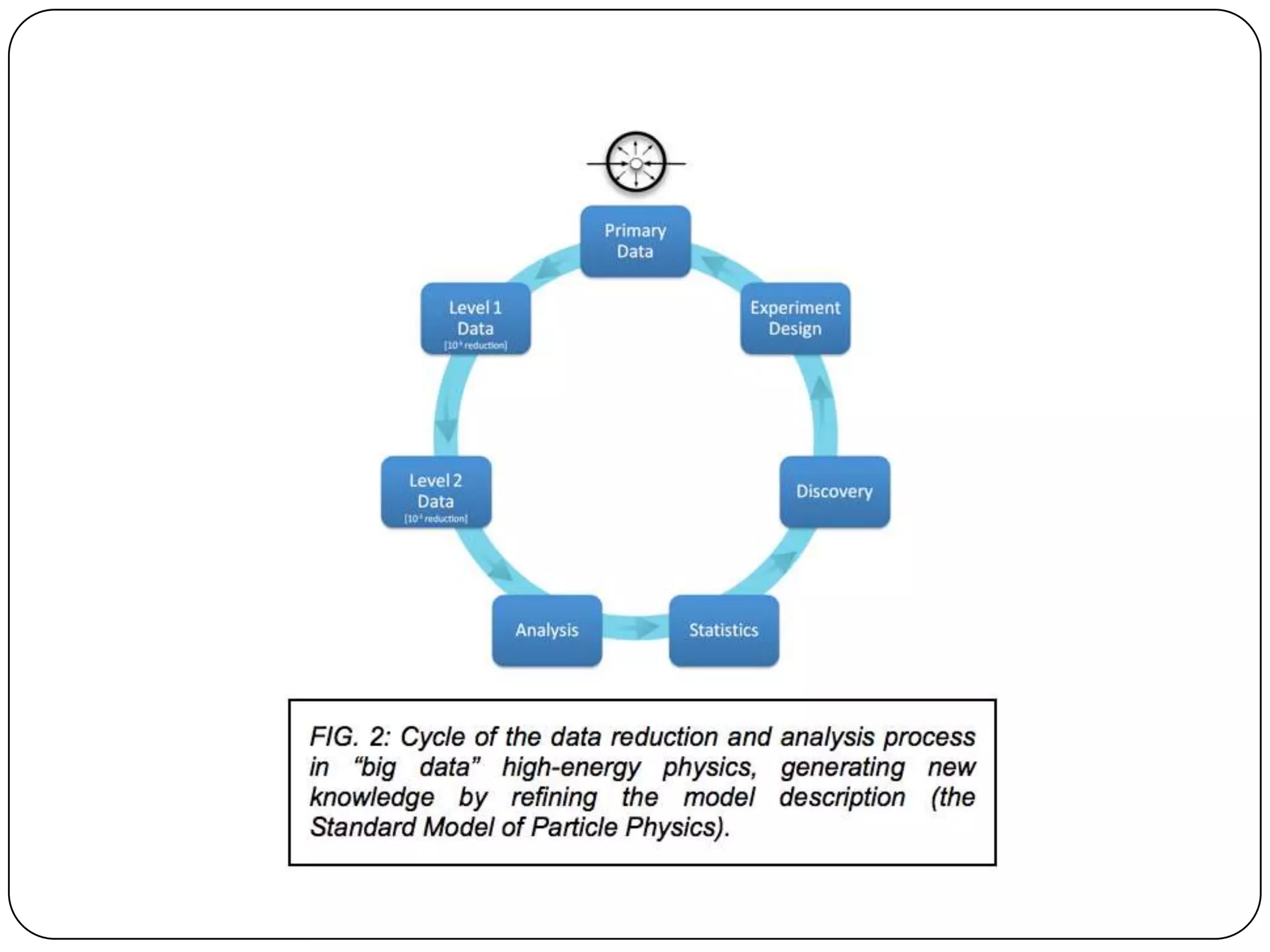
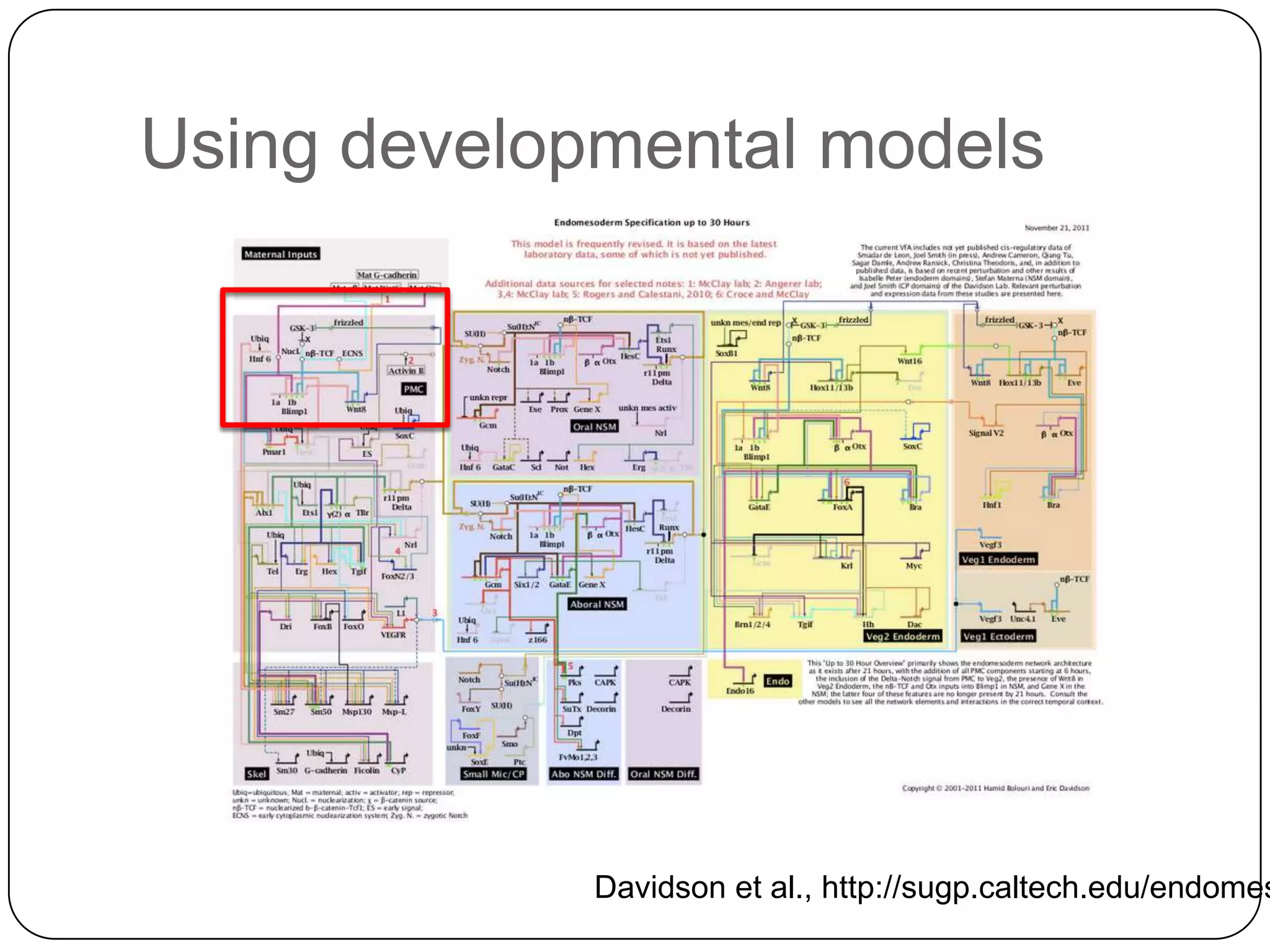
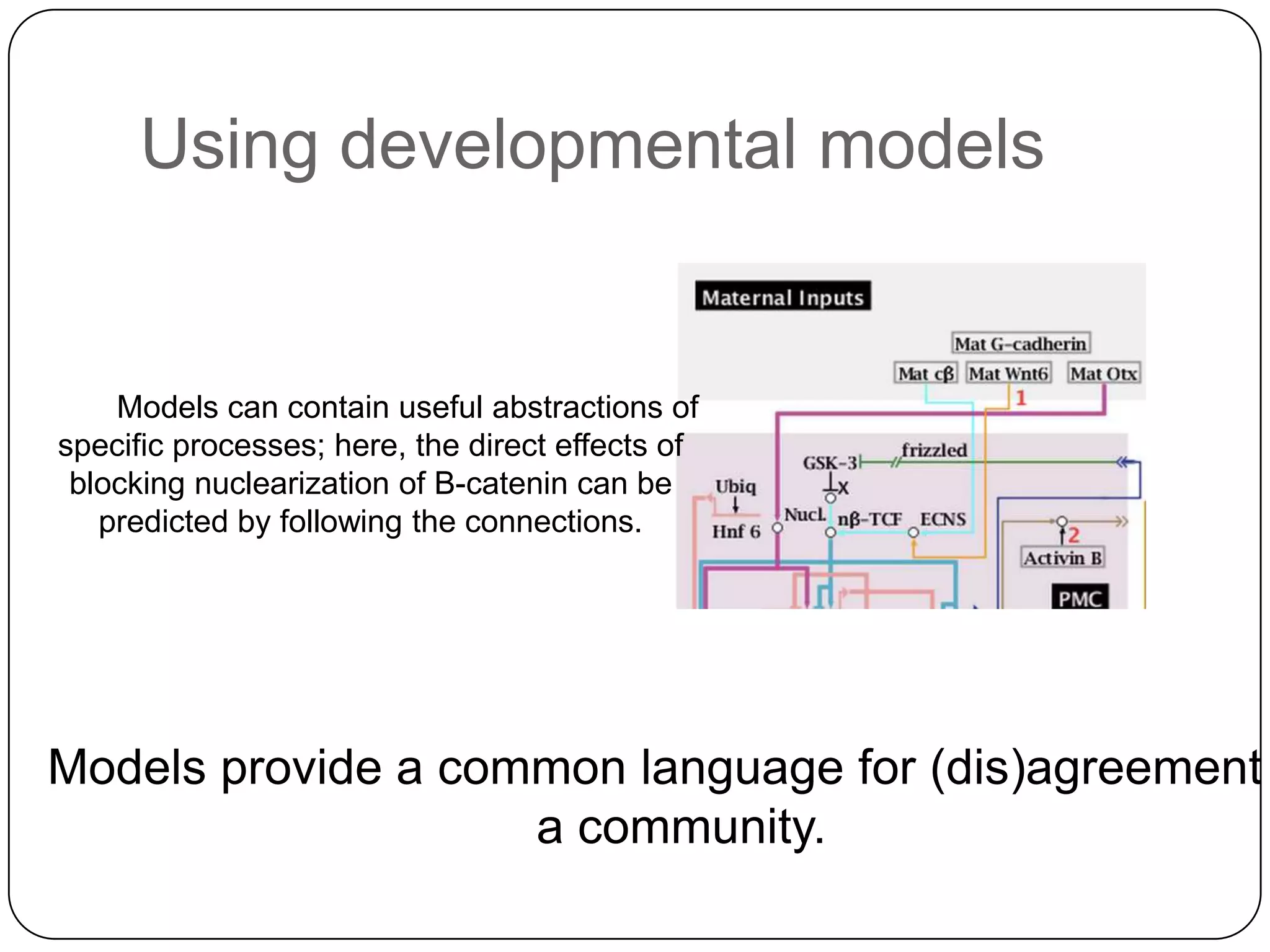
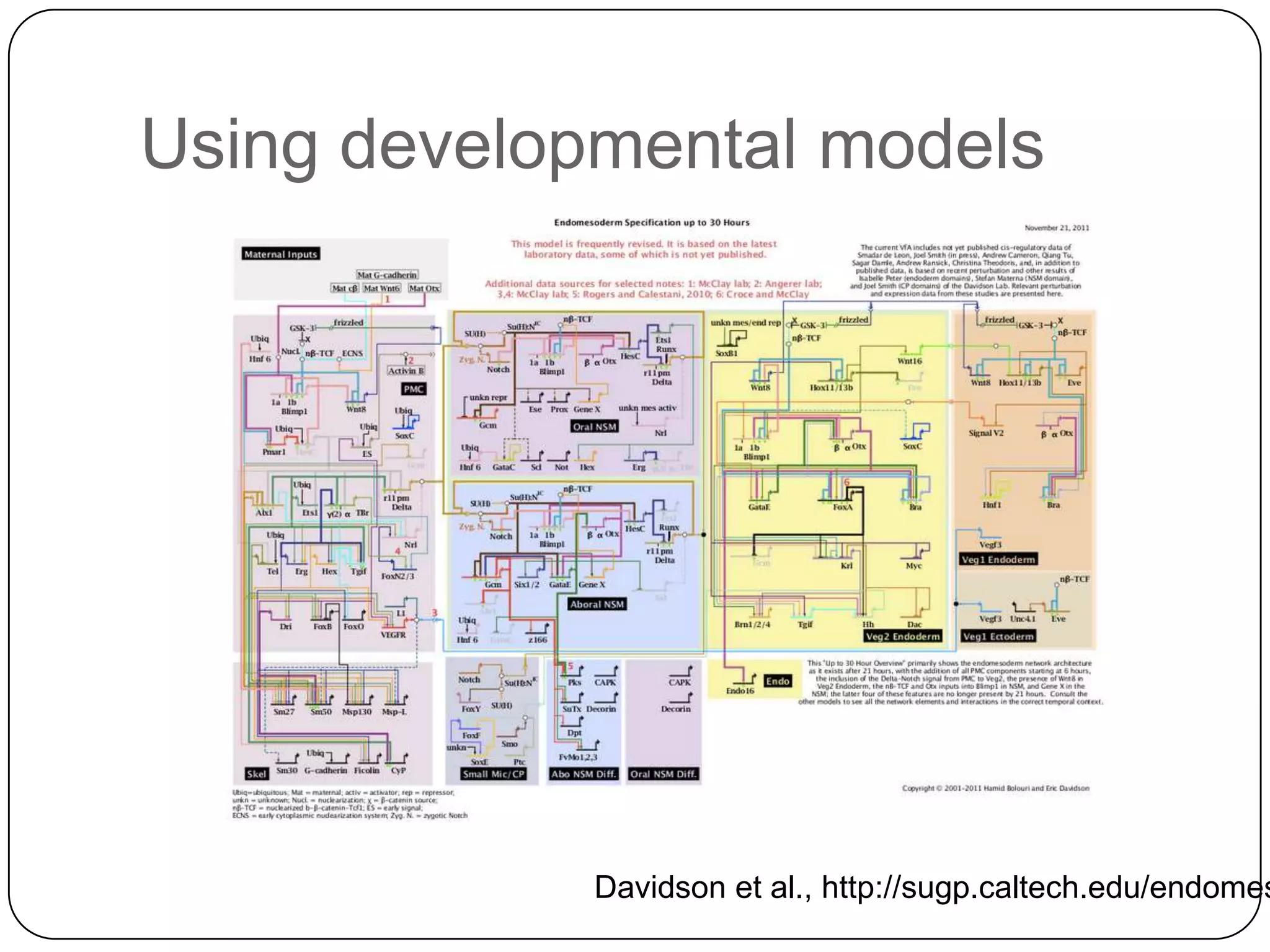
This document discusses challenges and opportunities for integrating large, heterogeneous biological data sets. It outlines the types of analysis and discovery that could be enabled, such as comparing data across studies. Technical challenges include incompatible identifiers and schemas between data sources. Common solutions attempt standardization but have limitations. The document examines Amazon's approach as a model, with principles like exposing all data through programmatic interfaces. It argues for a "platform" approach and combining data-driven and model-driven analysis to gain new insights. Developing services with end users in mind could help maximize data reuse.