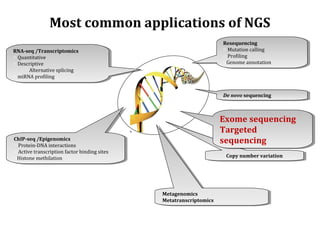
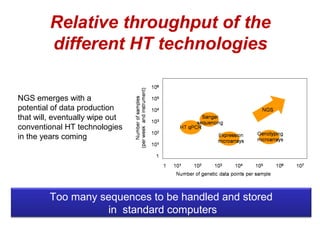
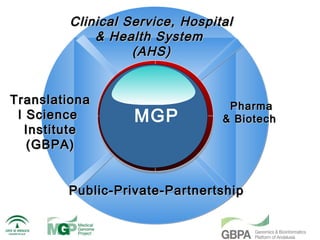
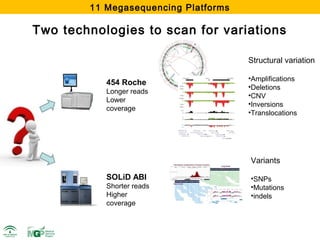
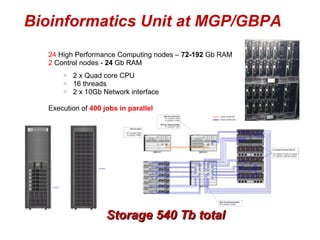
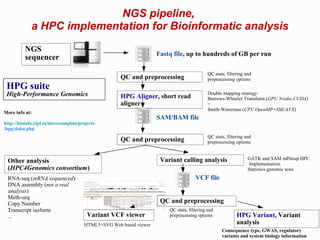
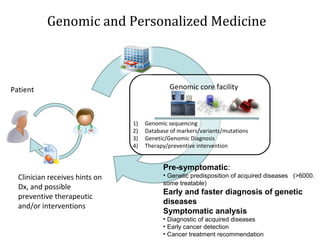
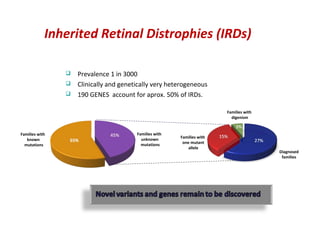
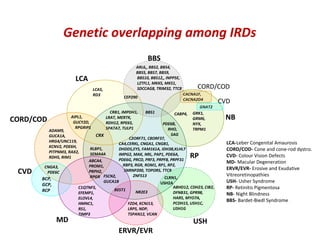
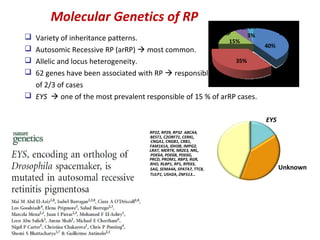
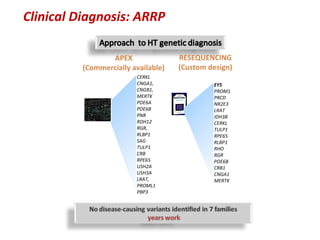
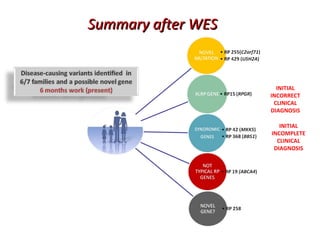
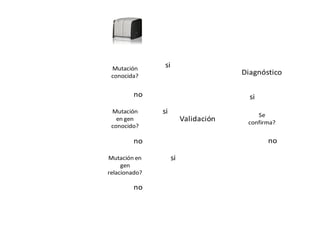
The document discusses the impact of next-generation sequencing (NGS) technology on genomic medicine, highlighting its ability to produce large datasets that enhance disease research and diagnosis. It outlines the challenges posed by big data in terms of processing, storage, and analysis while emphasizing the need for advanced computational technologies and collaborative efforts in bioinformatics. Furthermore, it highlights the establishment of resources and facilities, including a cloud-based platform, to facilitate genomic sequencing and improve healthcare outcomes.