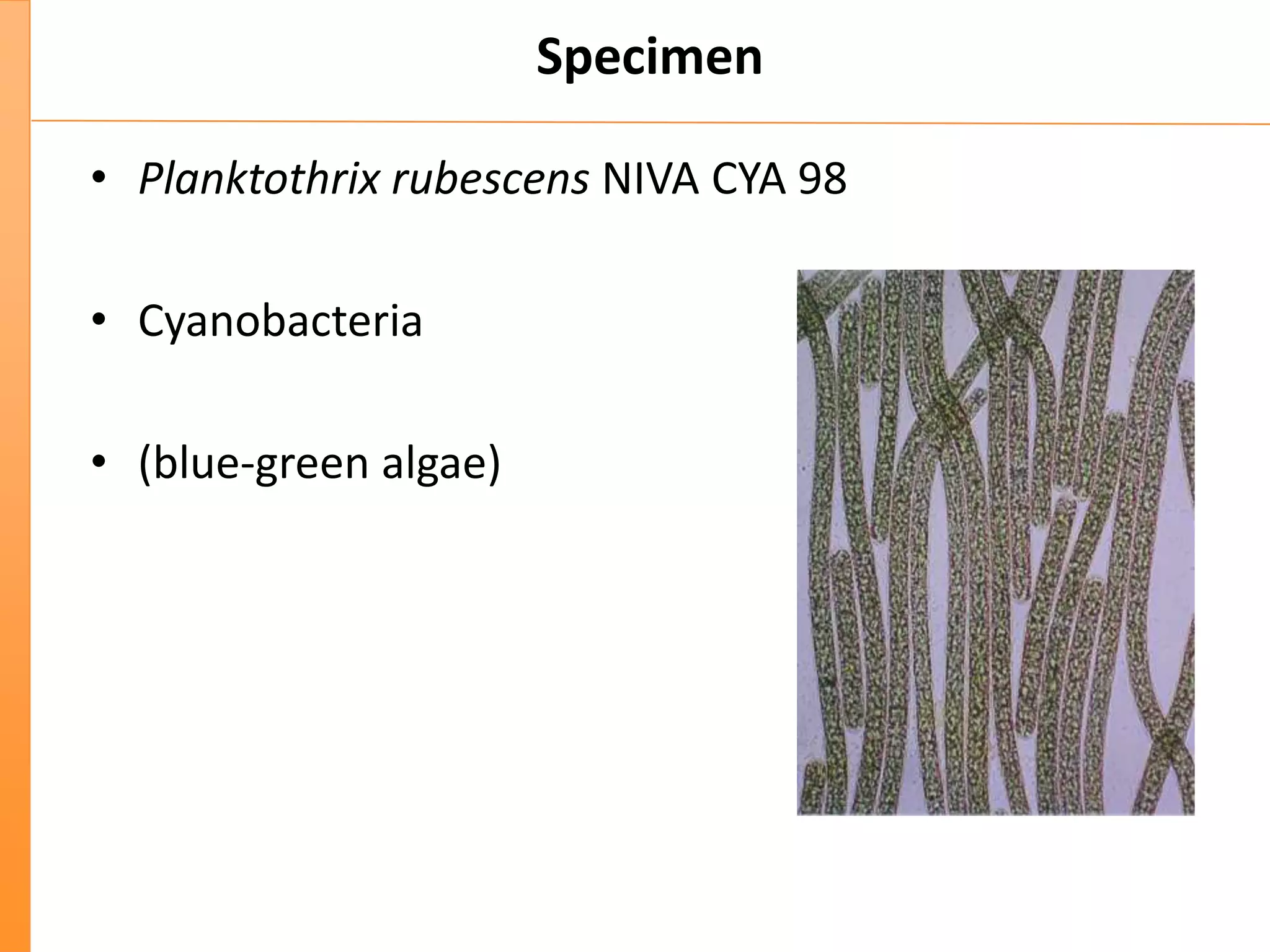
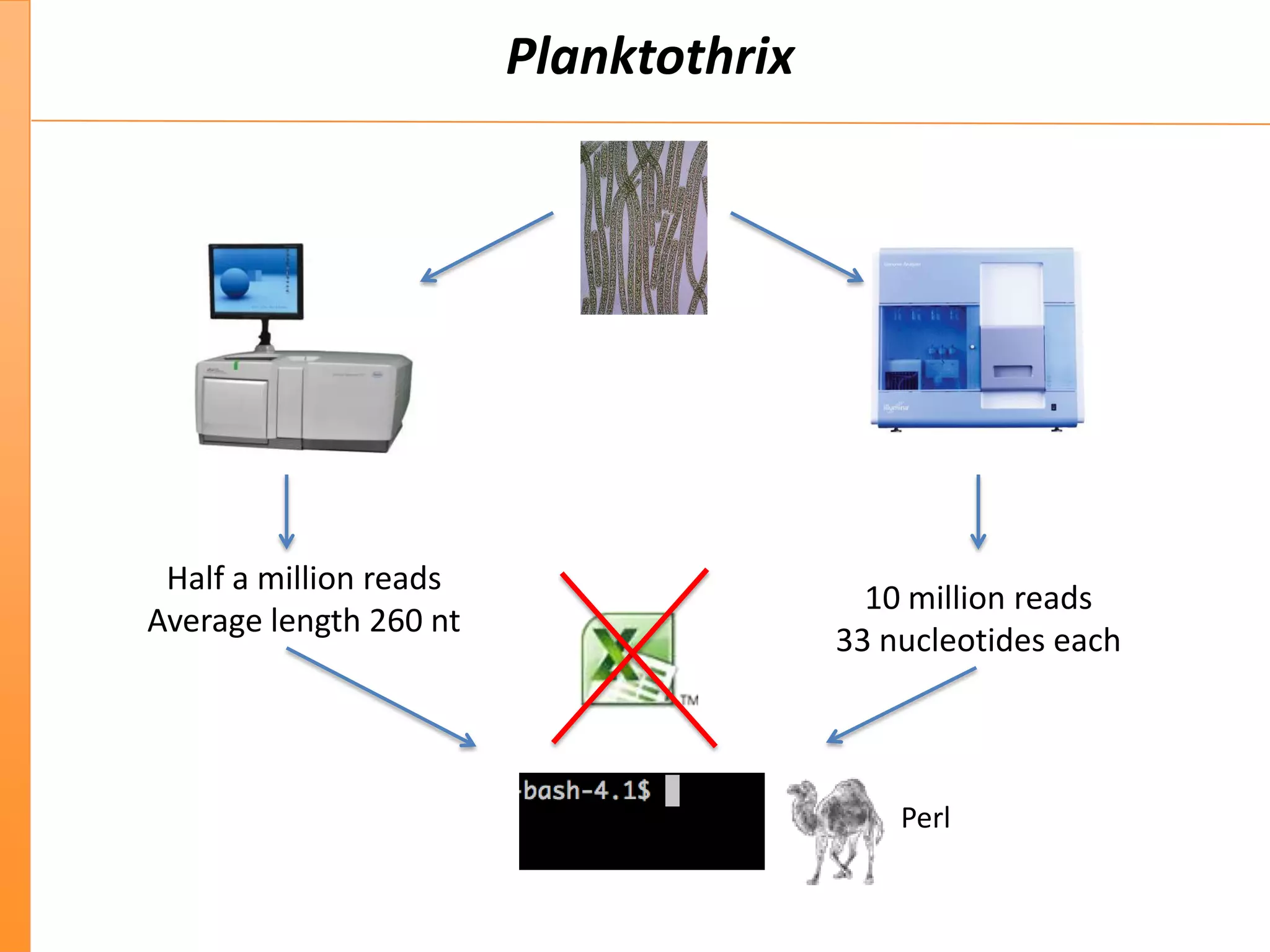
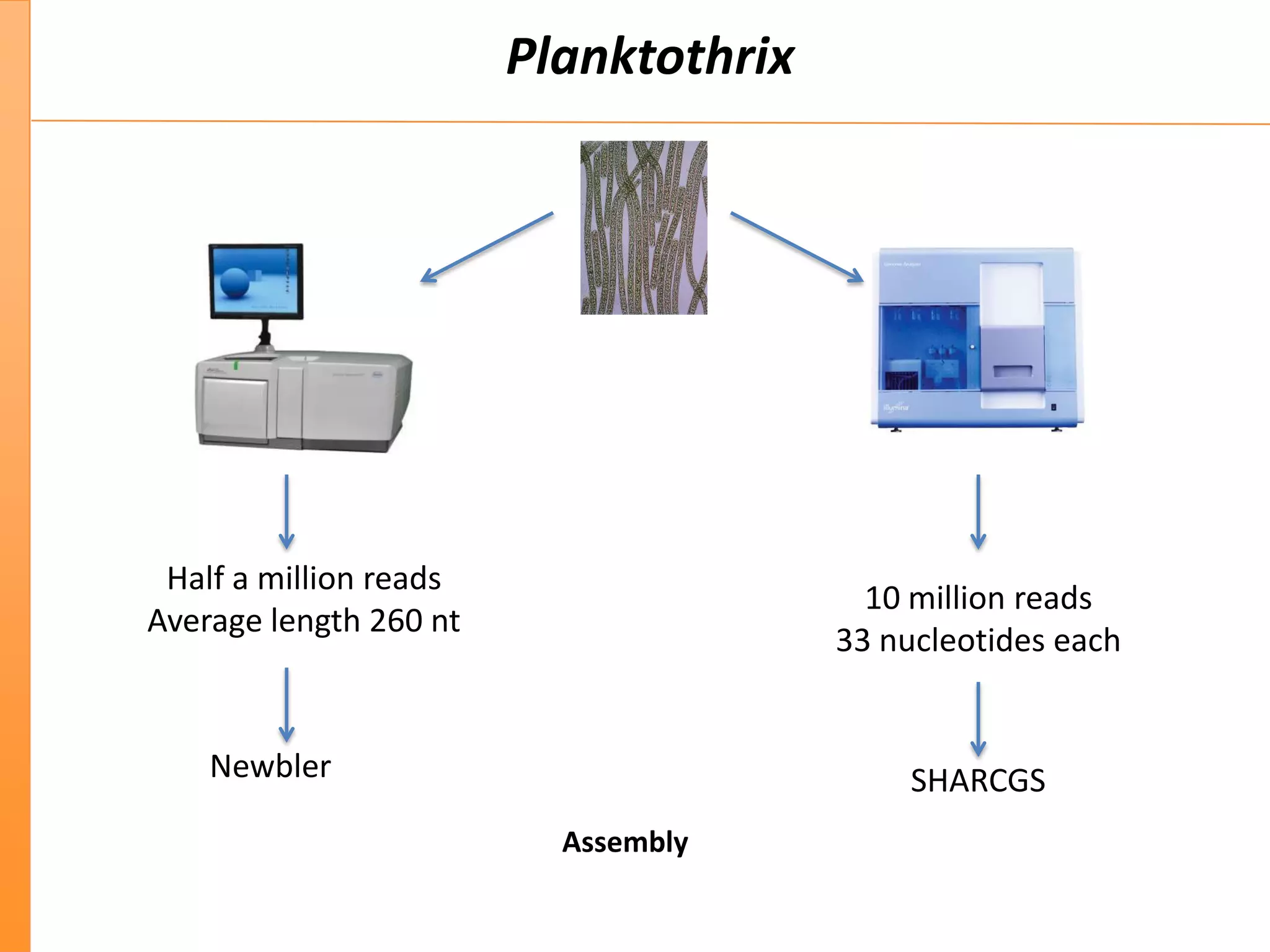
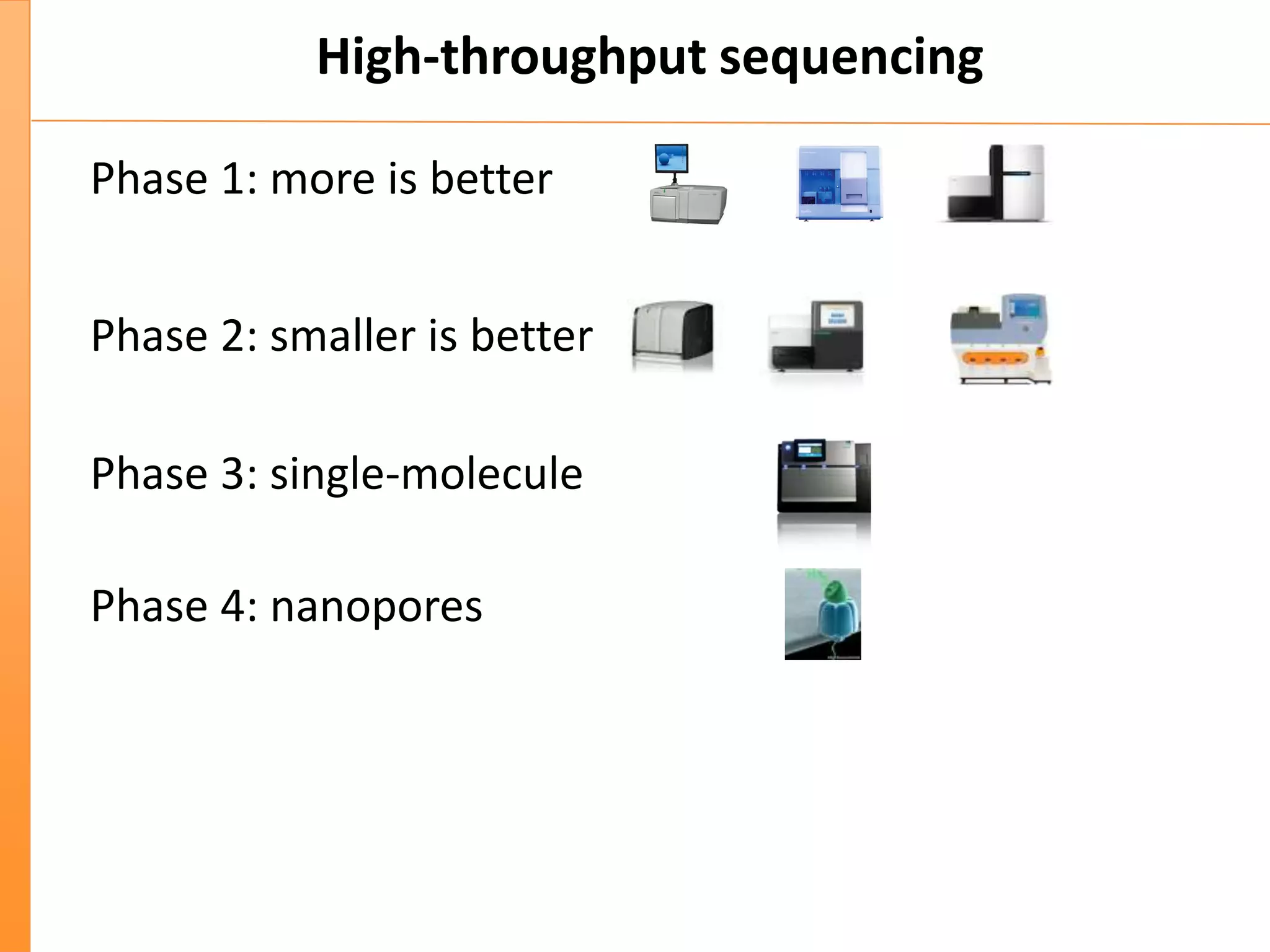
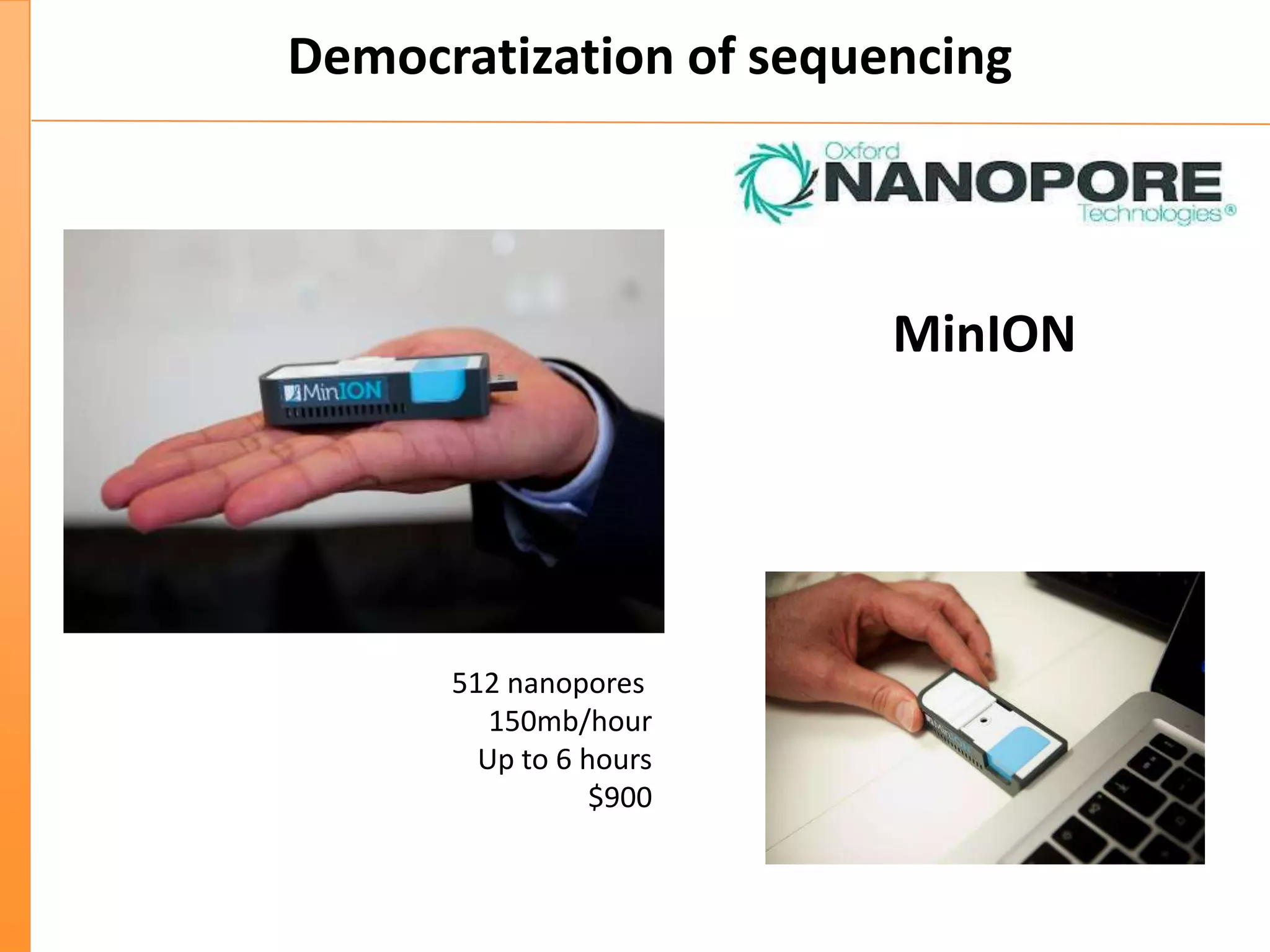
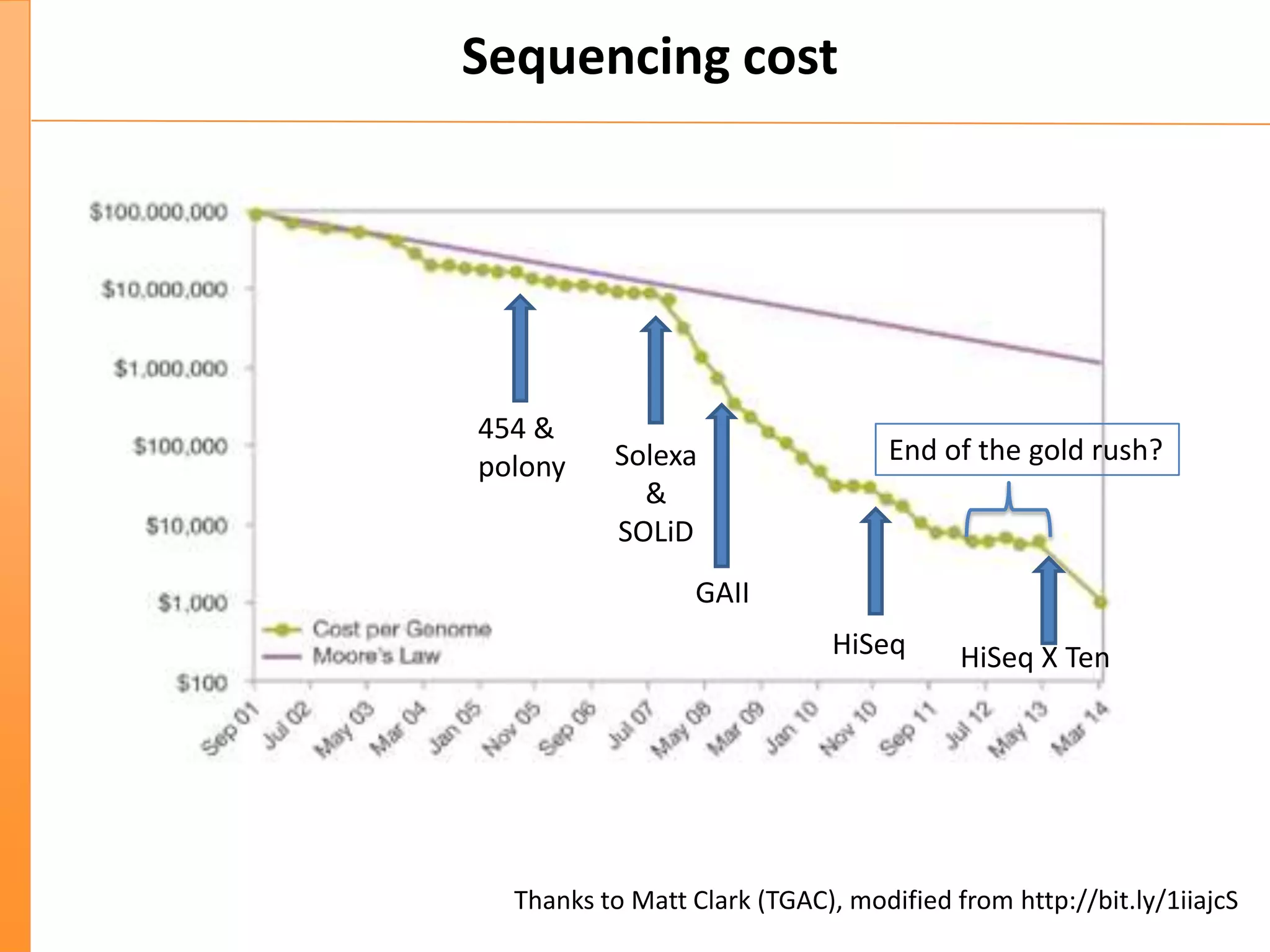
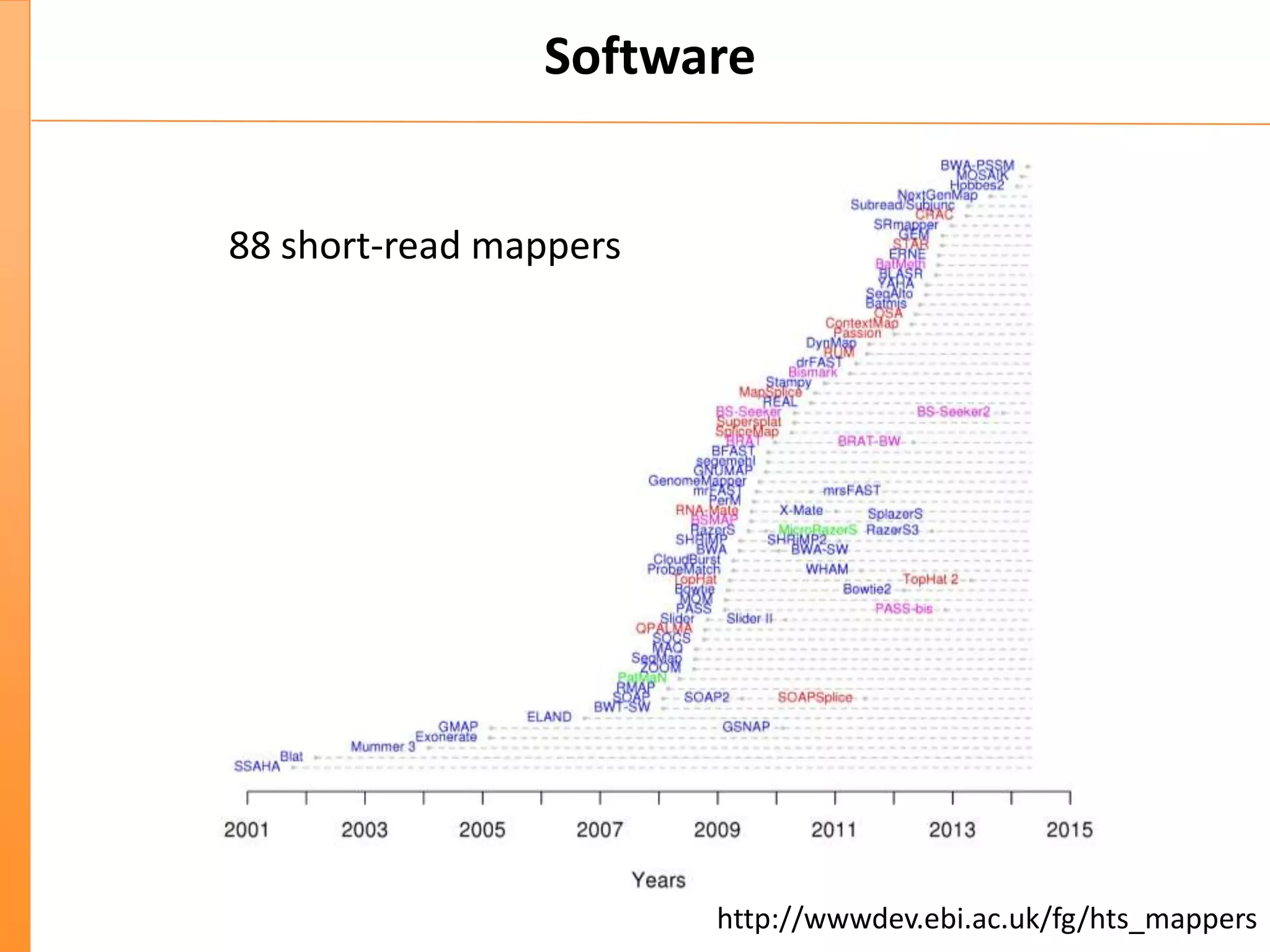
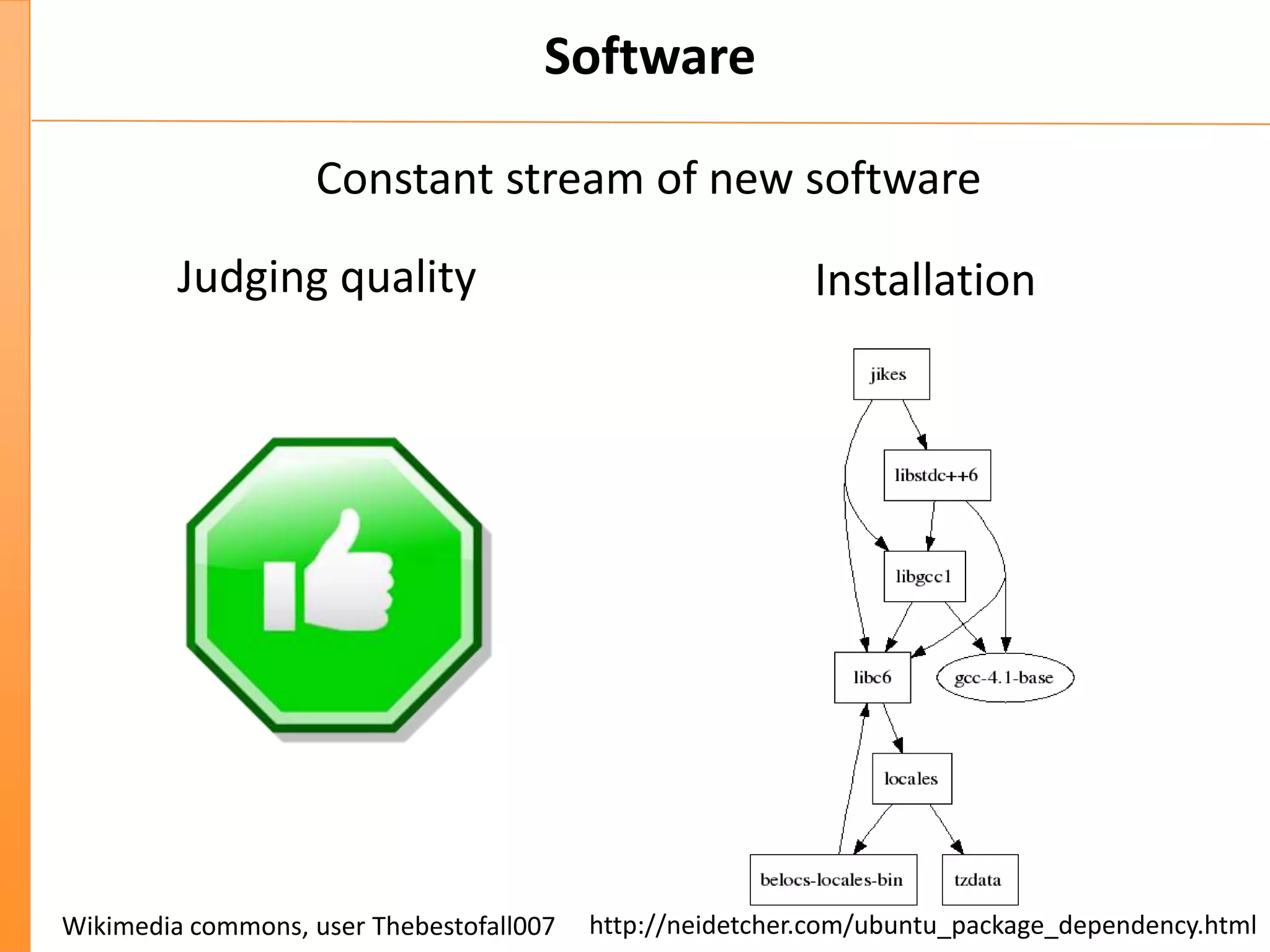
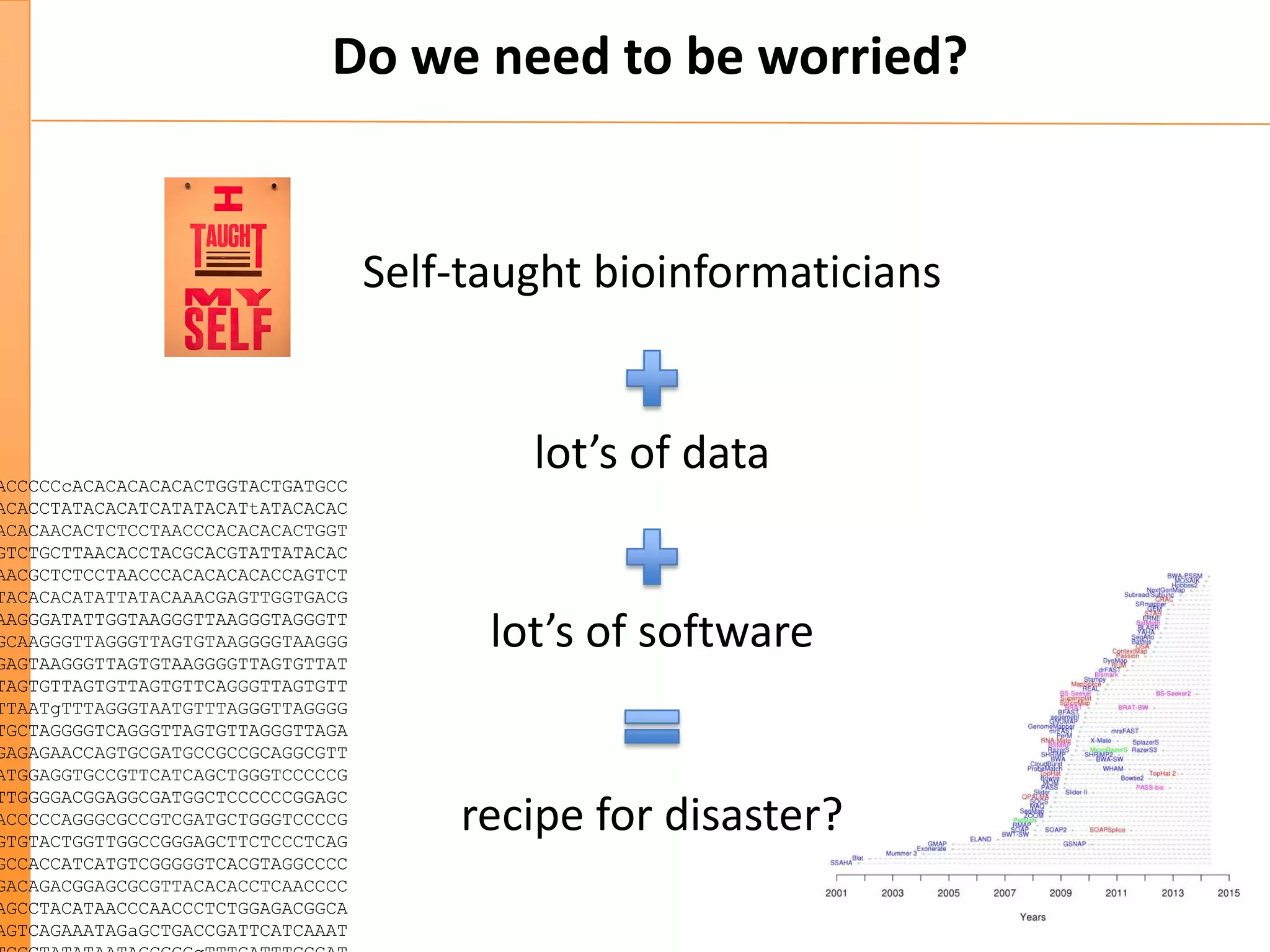
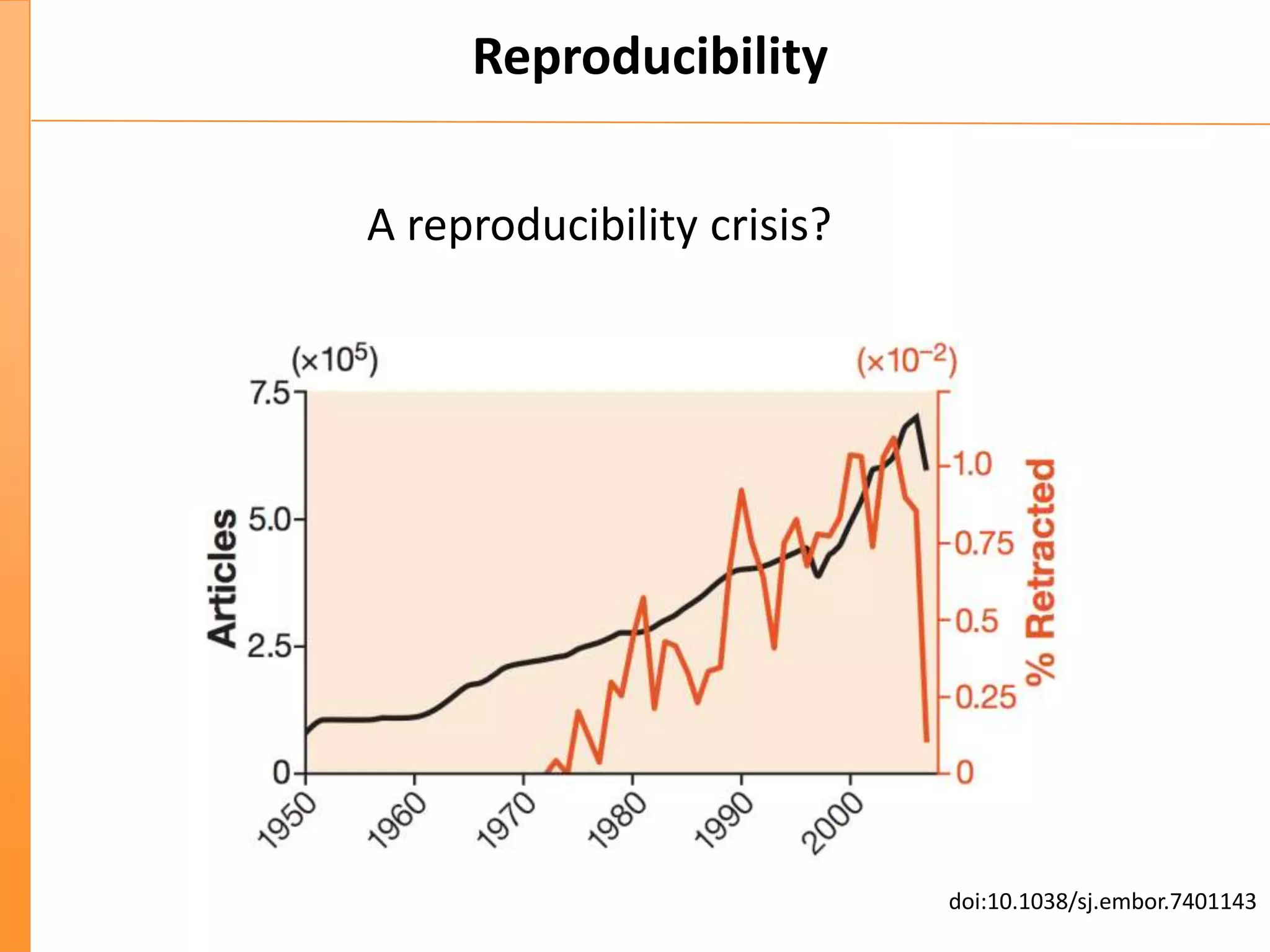
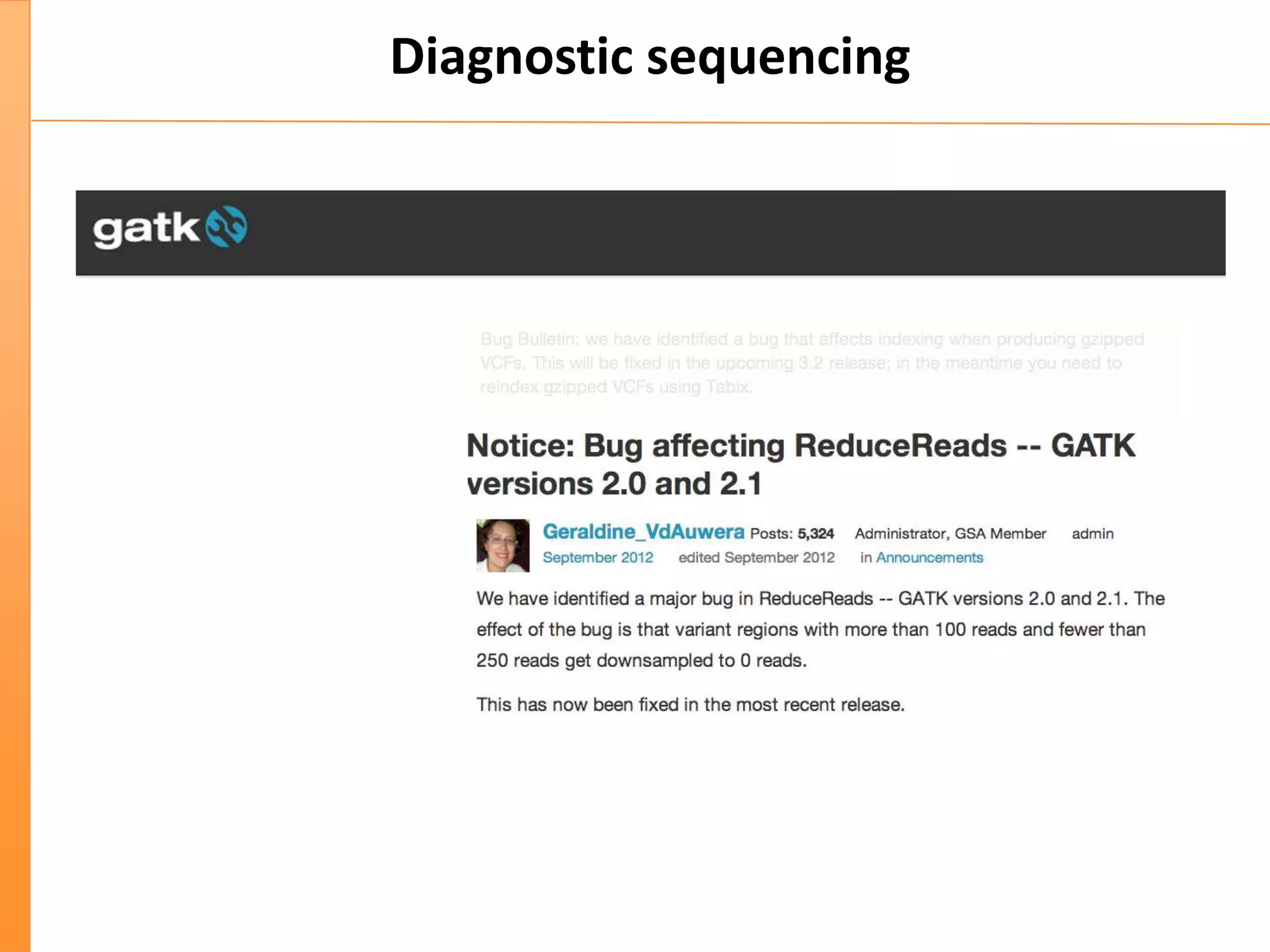
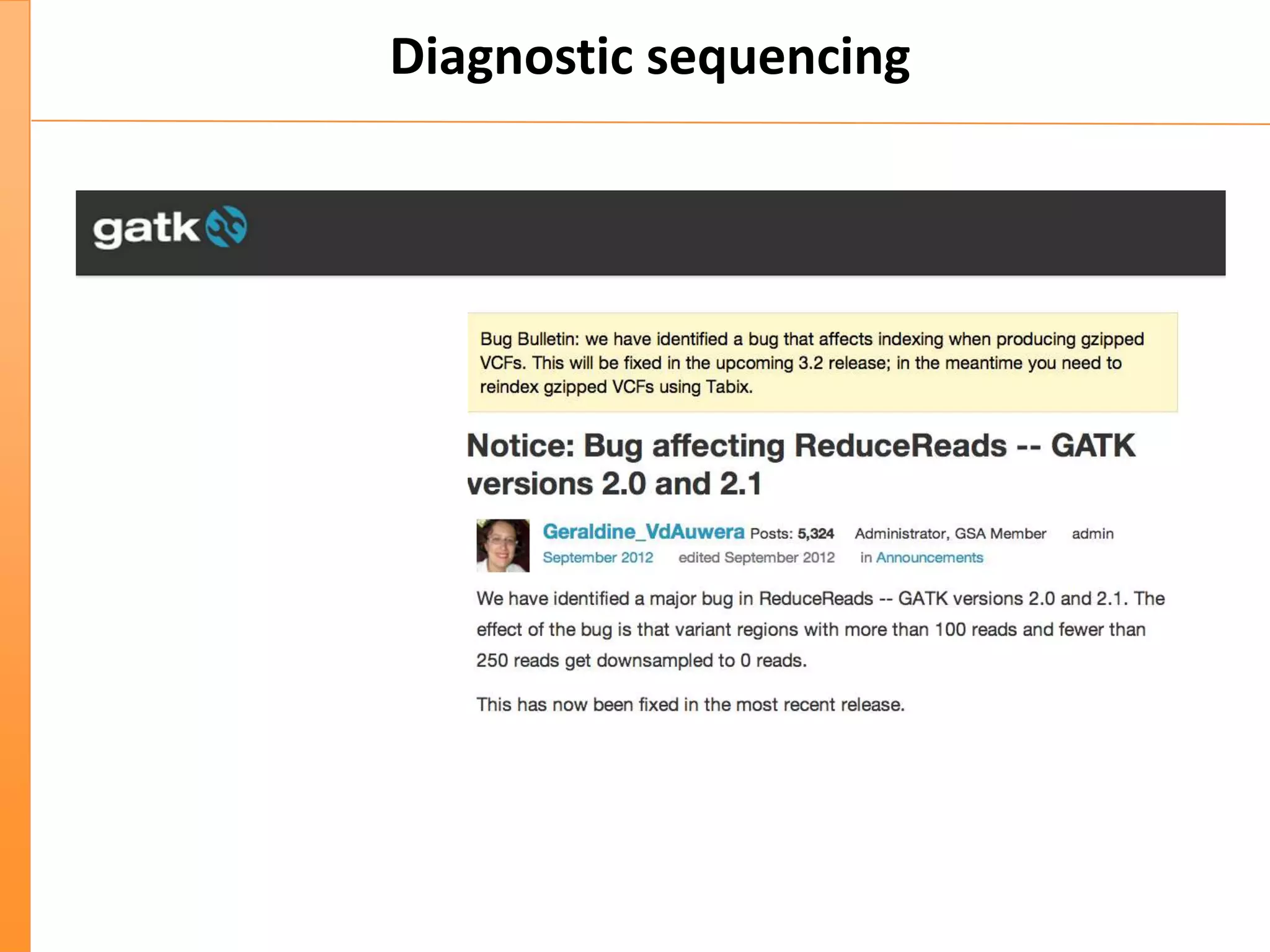
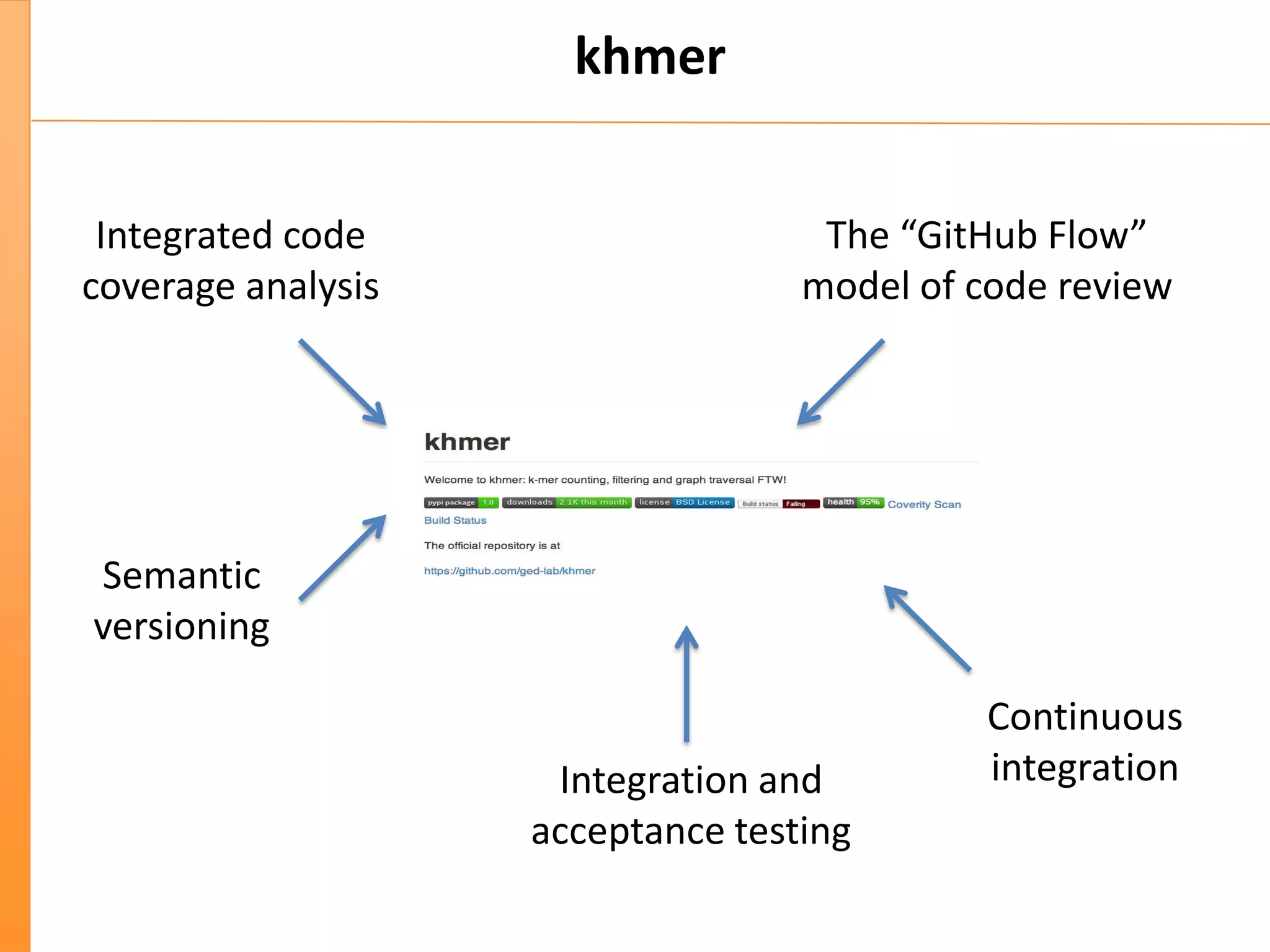
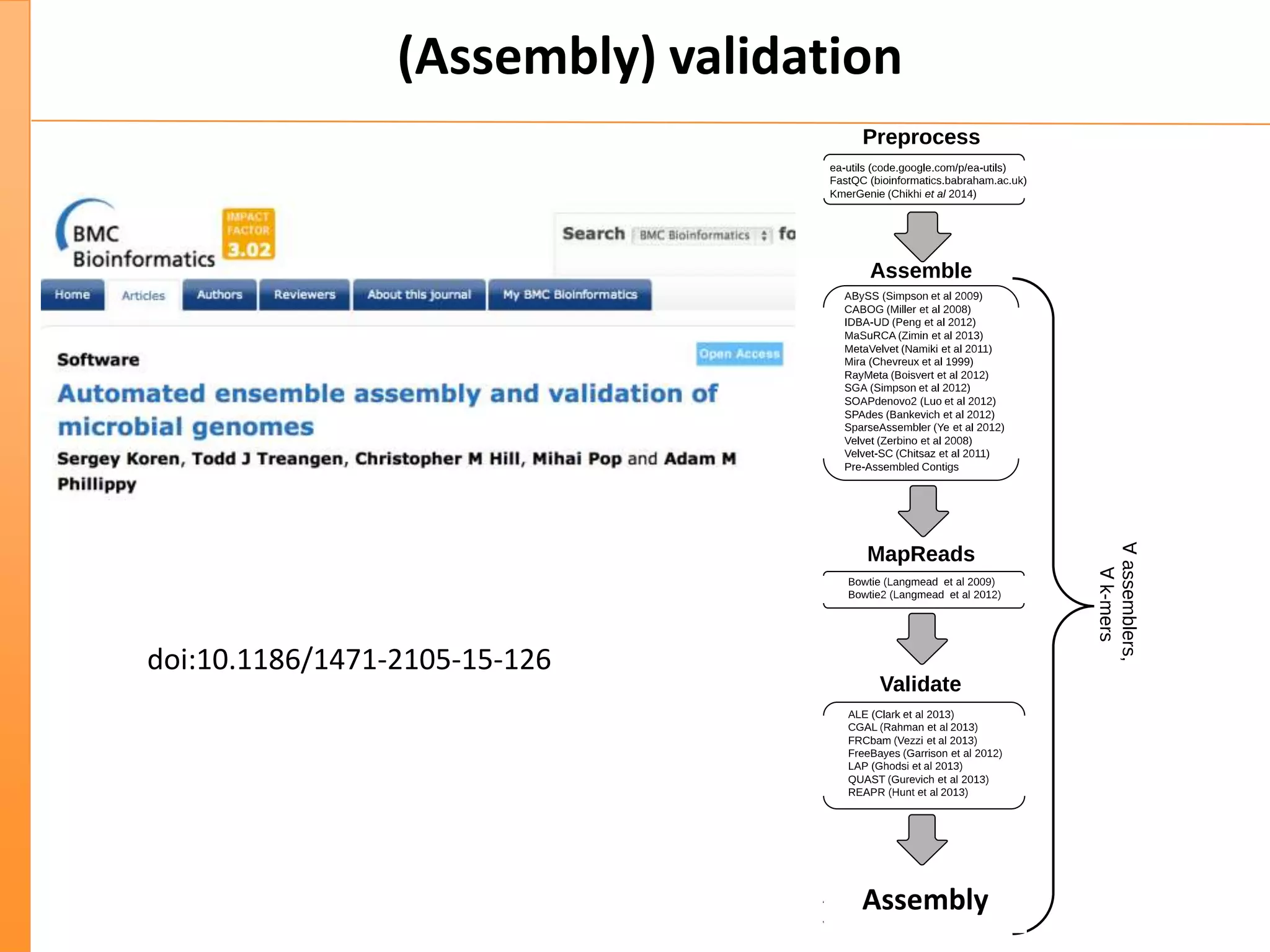
This document discusses the importance of best coding practices and reproducibility in programming for next-generation sequencing data analysis. It notes the large amount of data and numerous software tools now available pose challenges around obtaining correct and reproducible results. The document recommends following best practices like automated testing, version control, documentation, collaboration, and benchmarking tools to help address these challenges.