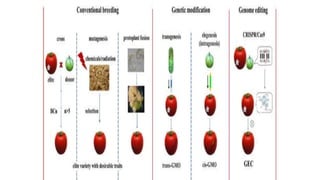
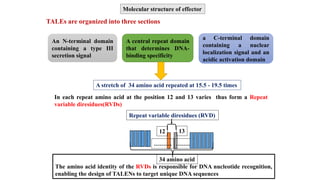
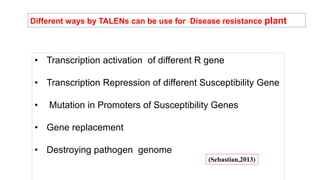
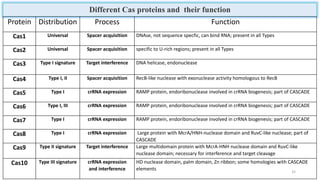
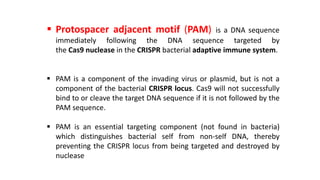
This document discusses genome editing techniques. It begins by defining genomes and how they consist of DNA or RNA that contains both coding and non-coding regions. It then discusses several methods of genome editing including zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and the CRISPR-Cas system. Each method uses engineered nucleases to introduce targeted double-strand breaks in DNA, allowing the cell's repair mechanisms to modify the genome. The CRISPR-Cas system was selected as the breakthrough of the year in 2015 due to its simplicity, efficiency and precision for genome editing applications.