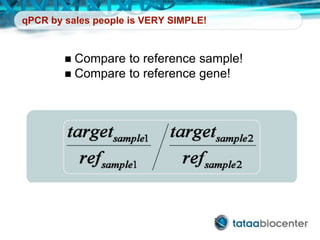
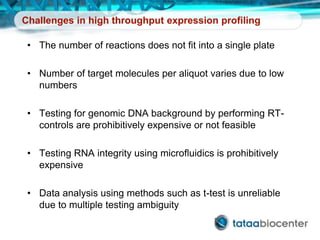
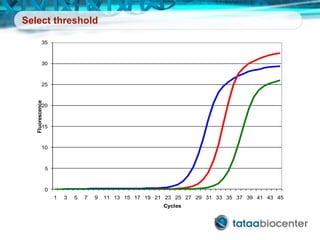
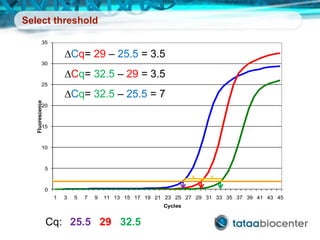
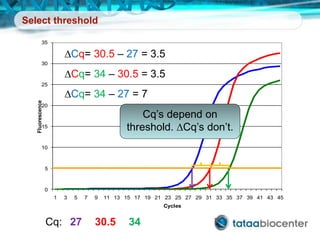
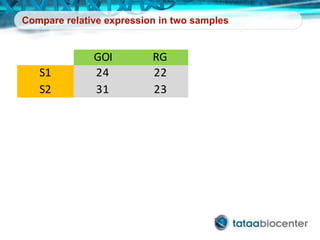
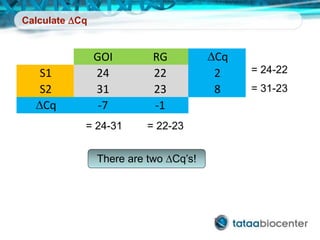
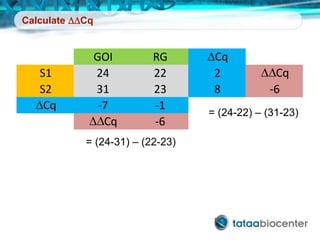
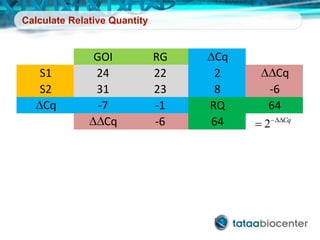
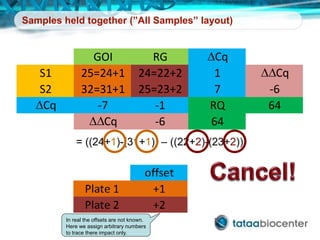
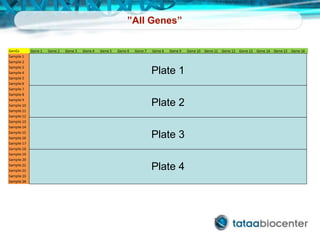
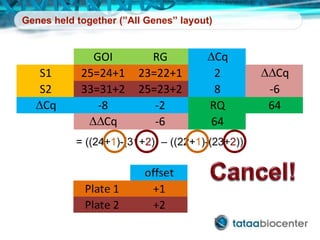
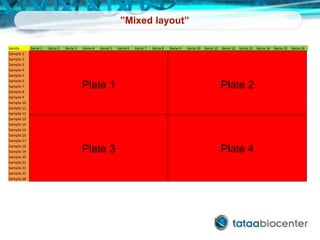
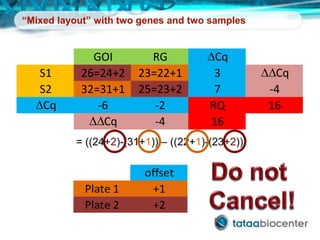
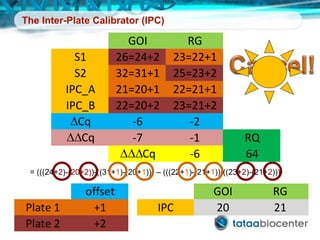
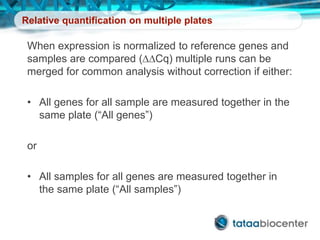
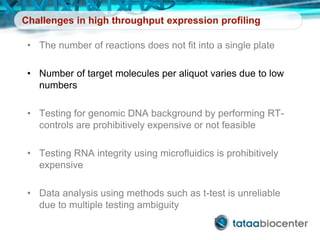
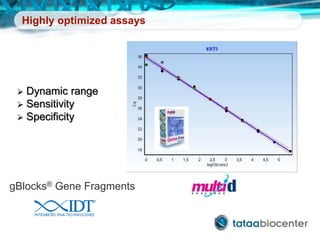
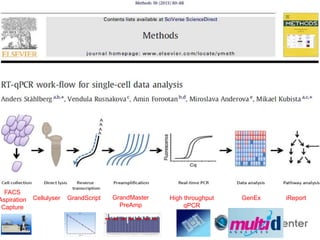
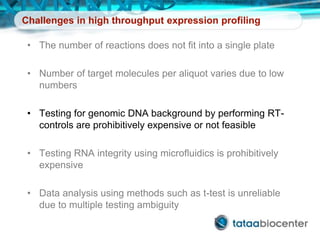
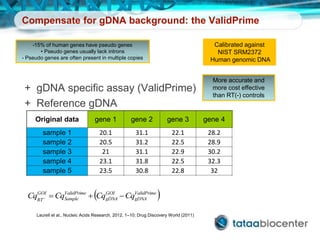
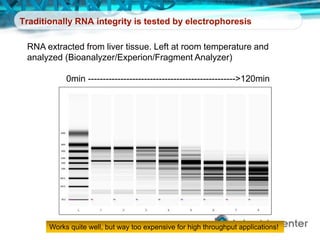
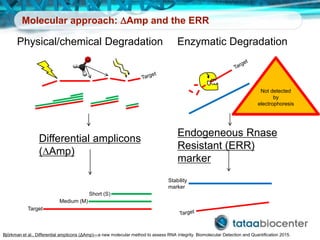
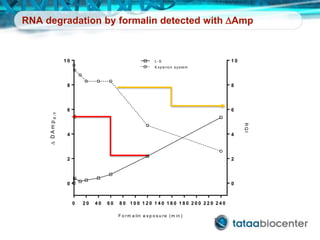
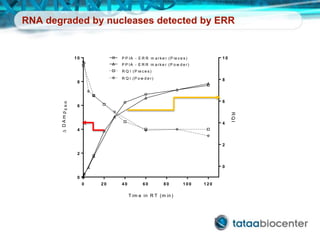
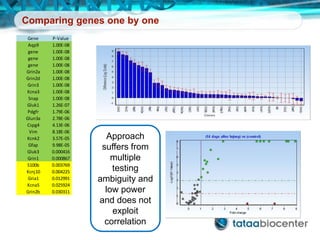
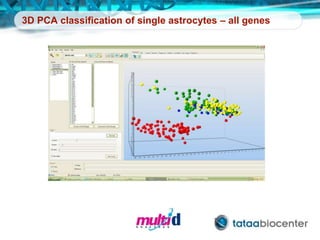
Dr. Mikael Kubista's presentation discusses the challenges of high-throughput qPCR, including issues with fitting reactions into a single plate, data analysis reliability, and the costs of necessary controls. It highlights the importance of interplate calibrators to manage variability and emphasizes the necessity for accurate RNA integrity testing. The document also outlines potential methodologies and tools for optimizing gene expression profiling in complex biological samples.