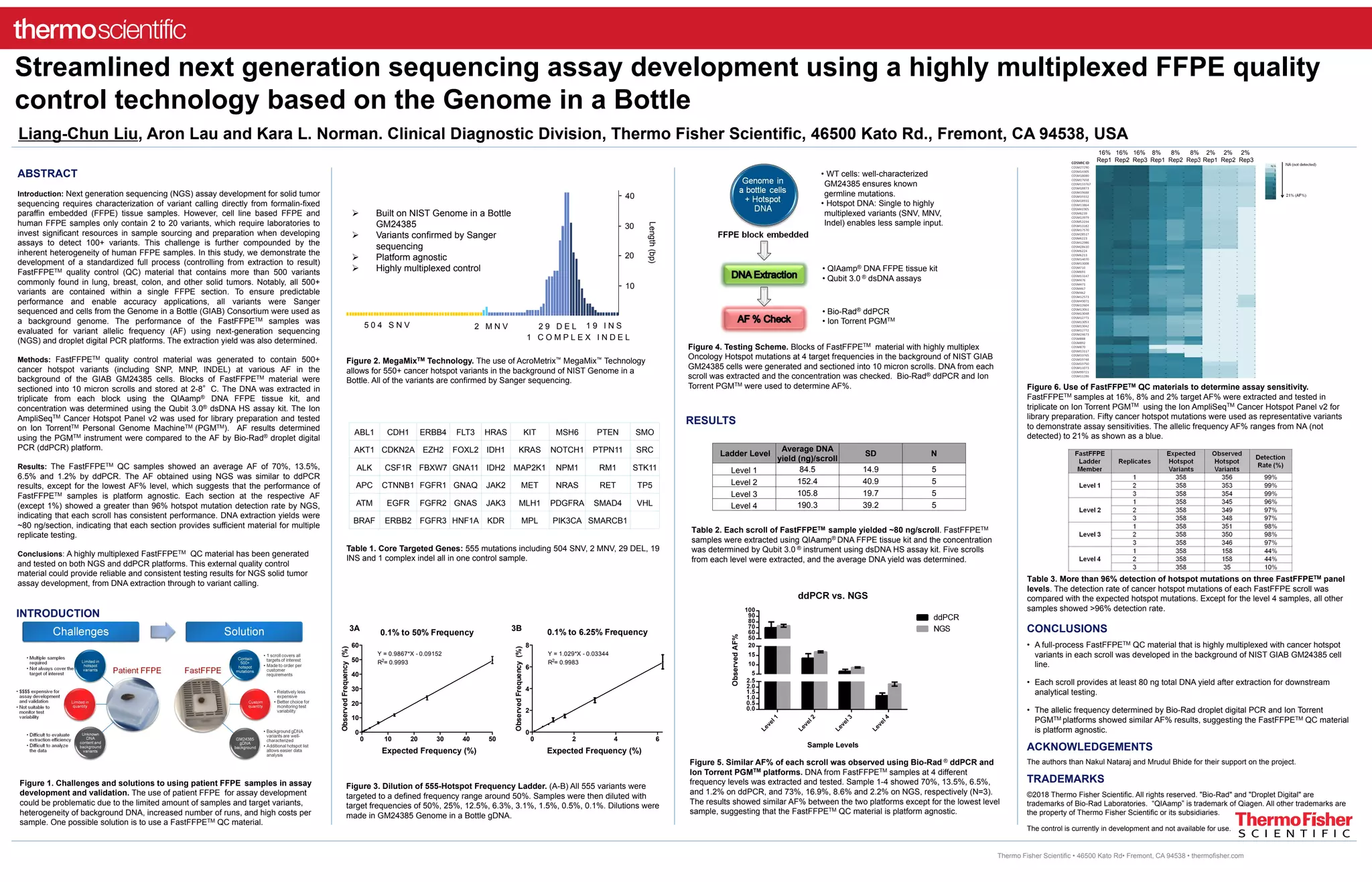
The document describes the development of a standardized quality control material, fastFFPE™, containing over 500 cancer hotspot variants for solid tumor sequencing, which is platform agnostic. Testing of extracted DNA showed similar variant allelic frequencies when compared between next generation sequencing (NGS) and droplet digital PCR (ddPCR). This fastFFPE QC material aims to enhance reliability and consistency in assay development, providing enough material for multiple tests with high detection rates.