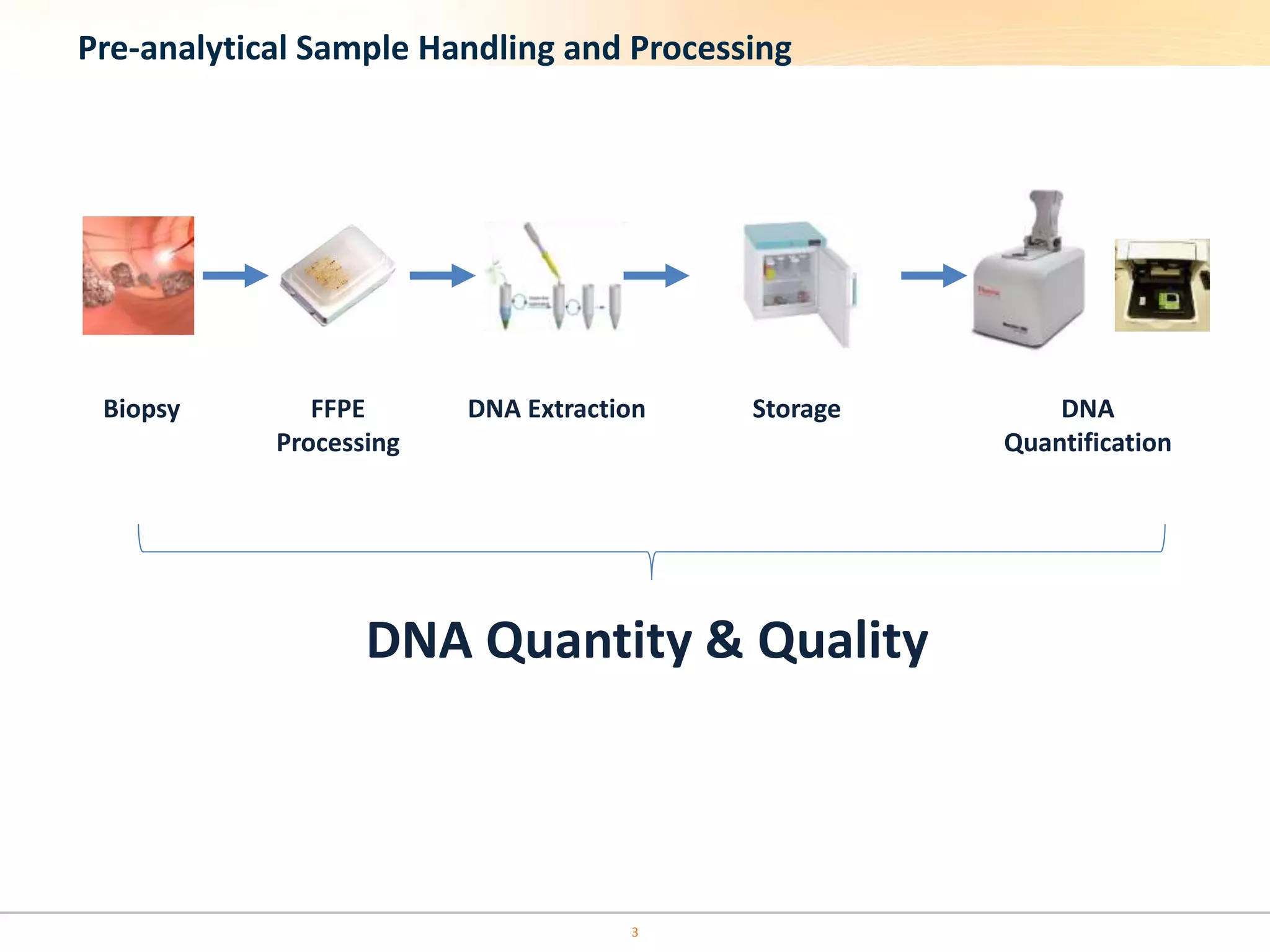
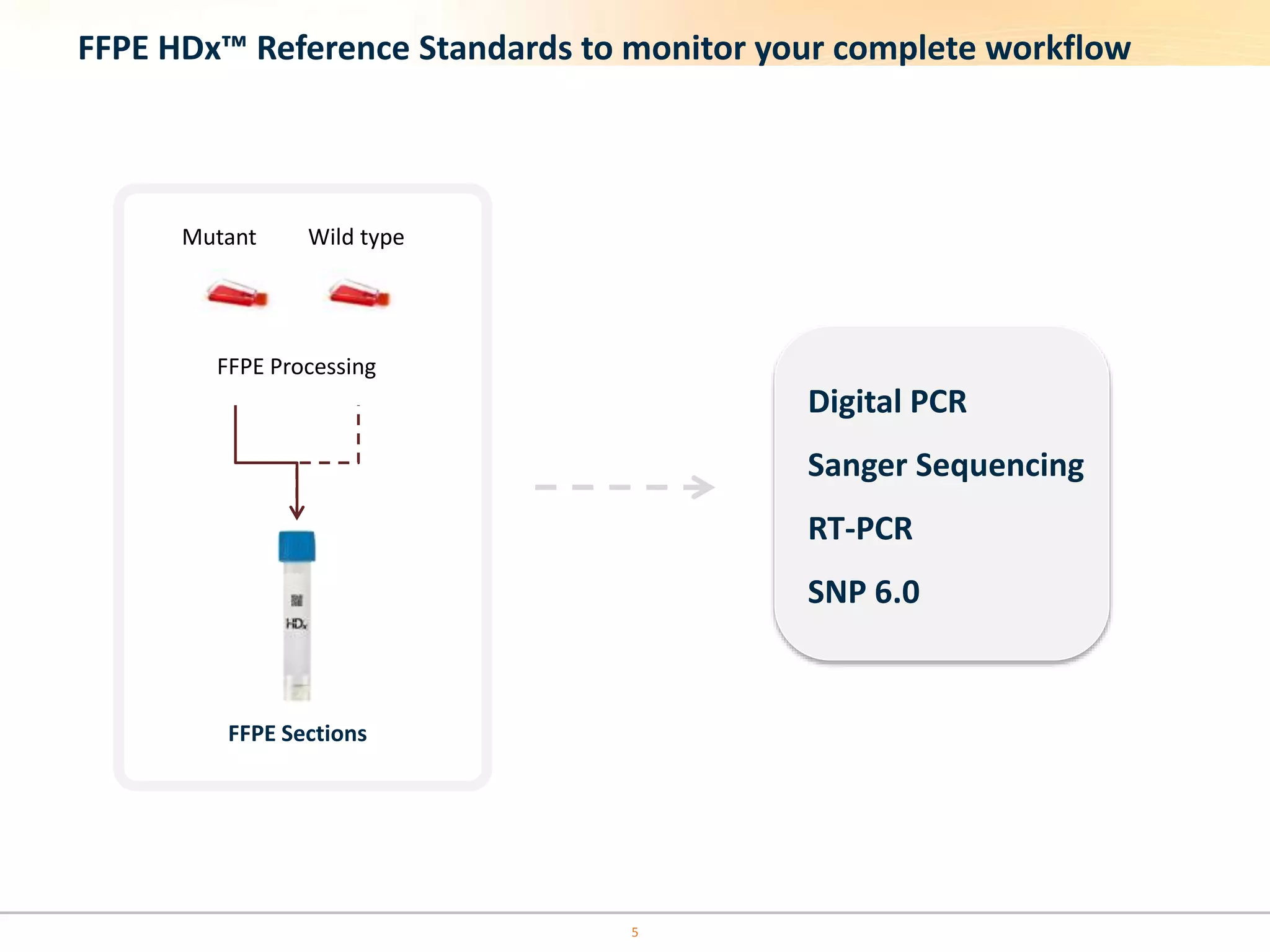
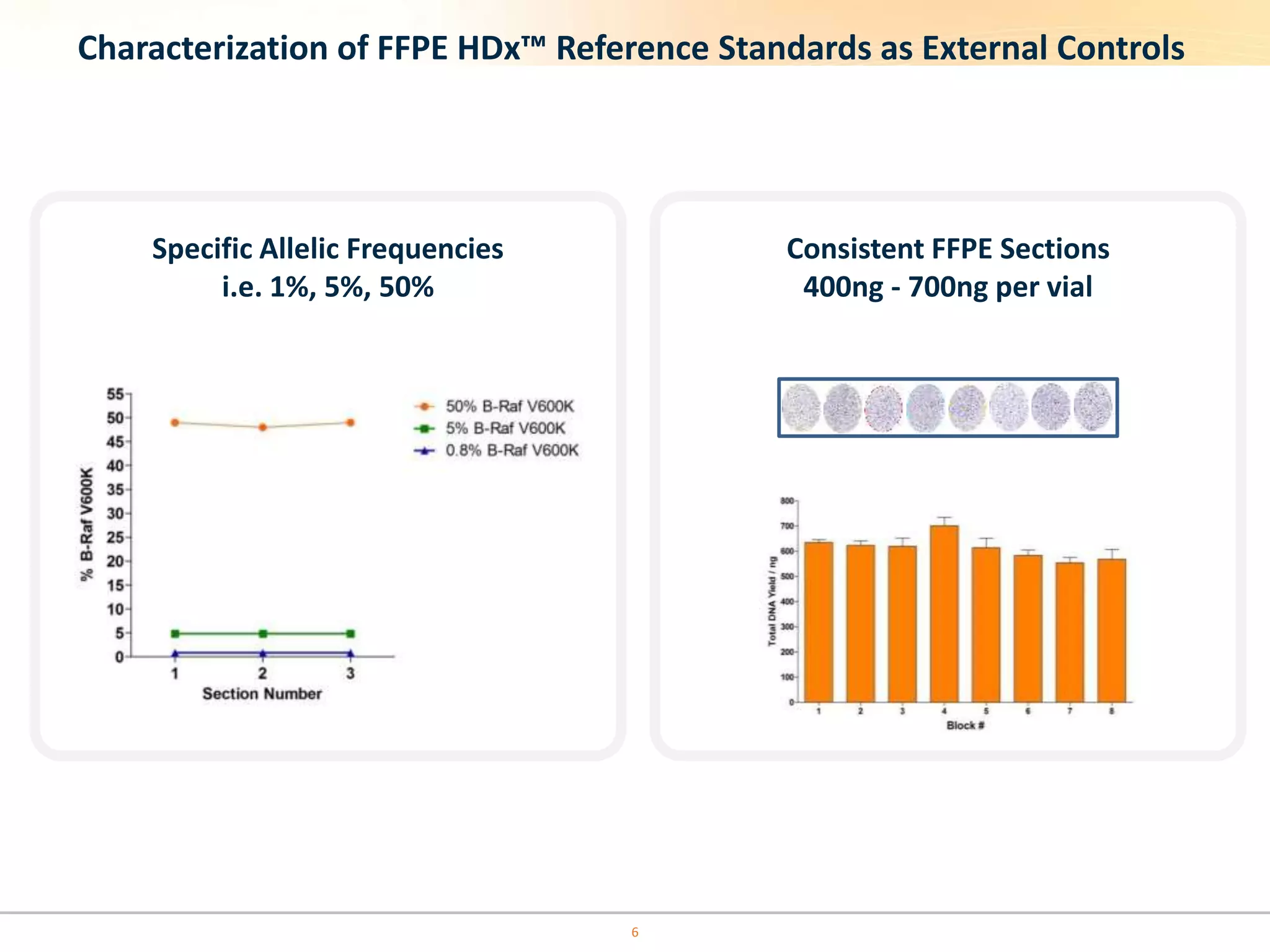
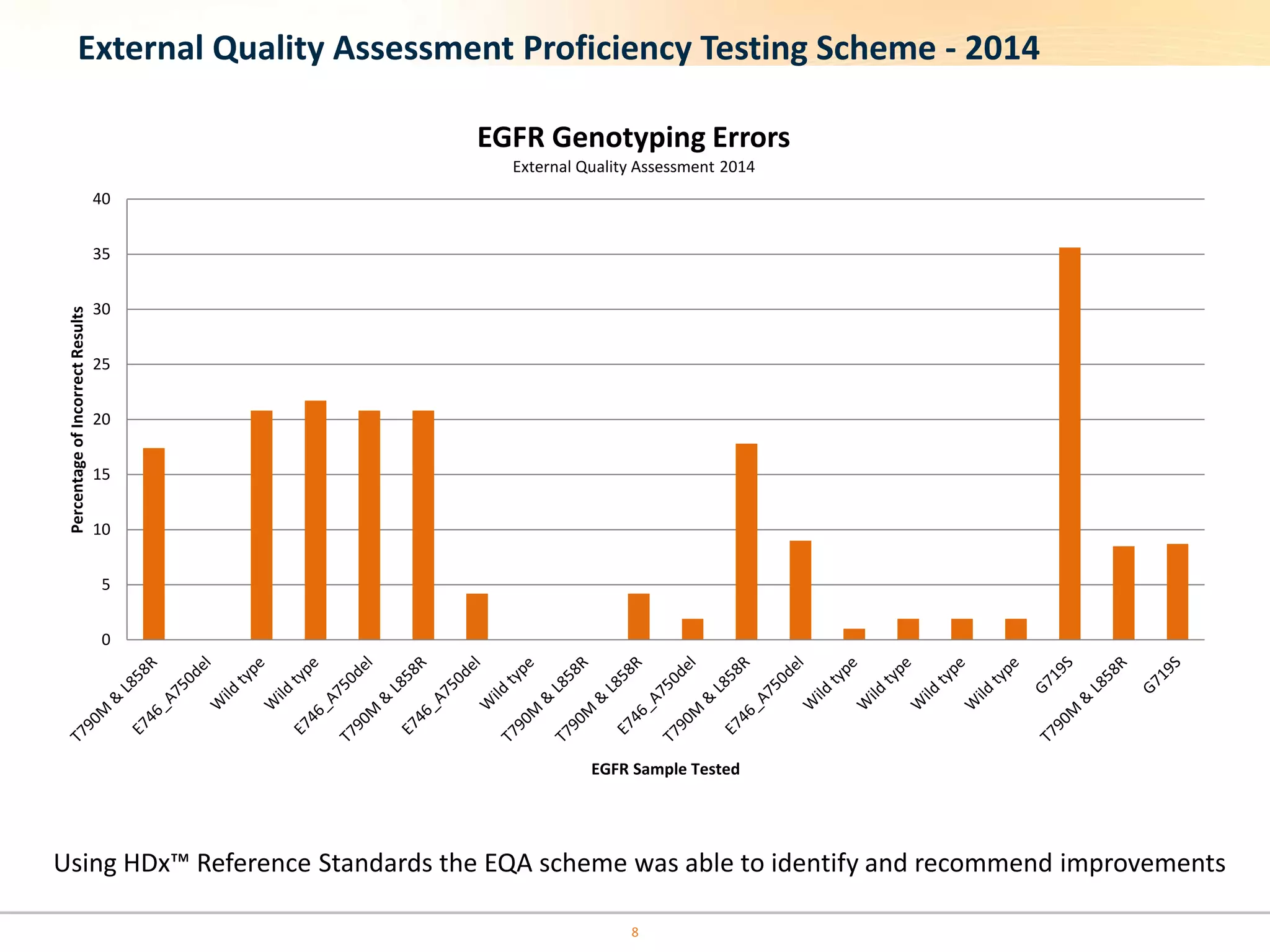
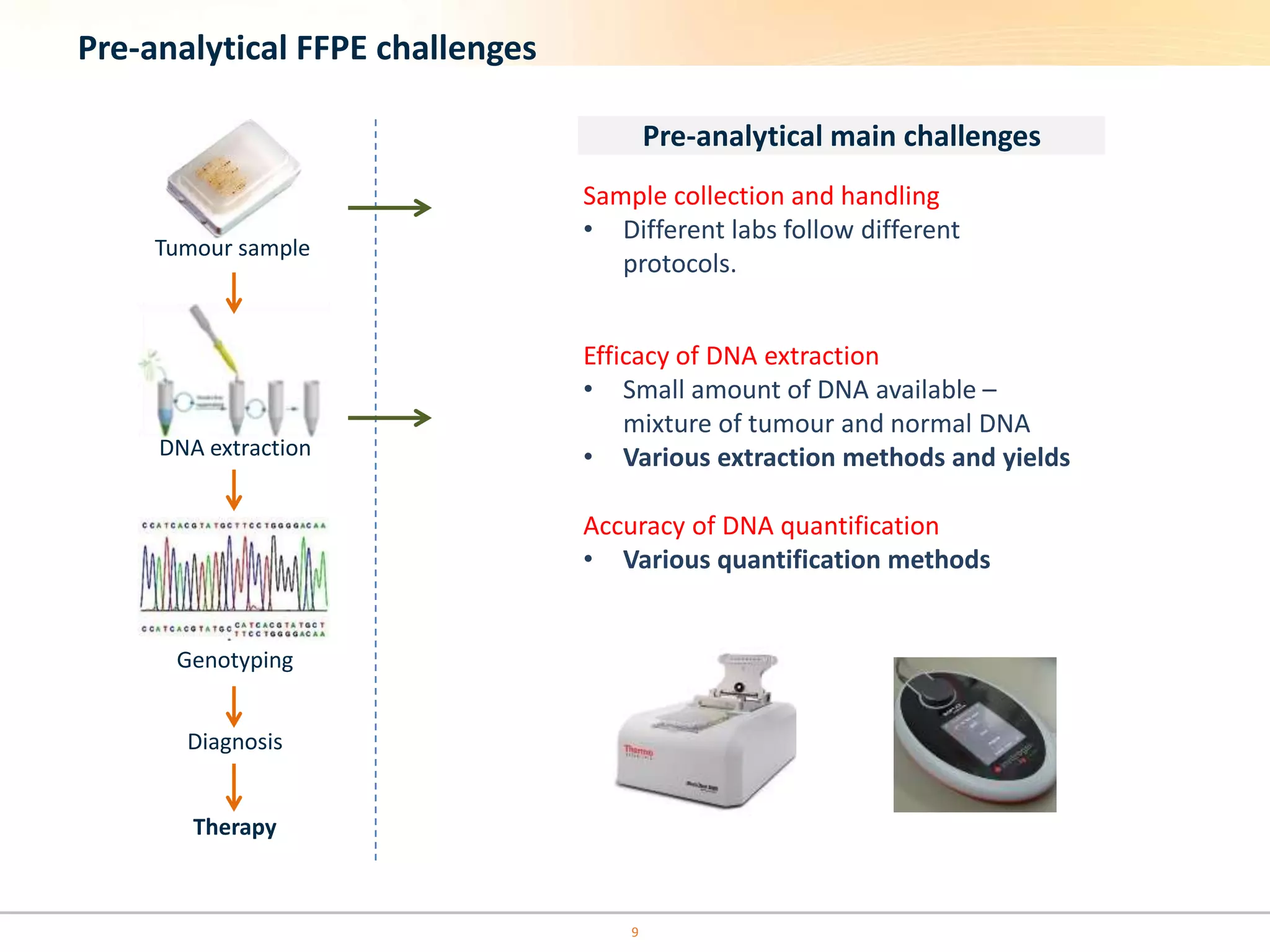
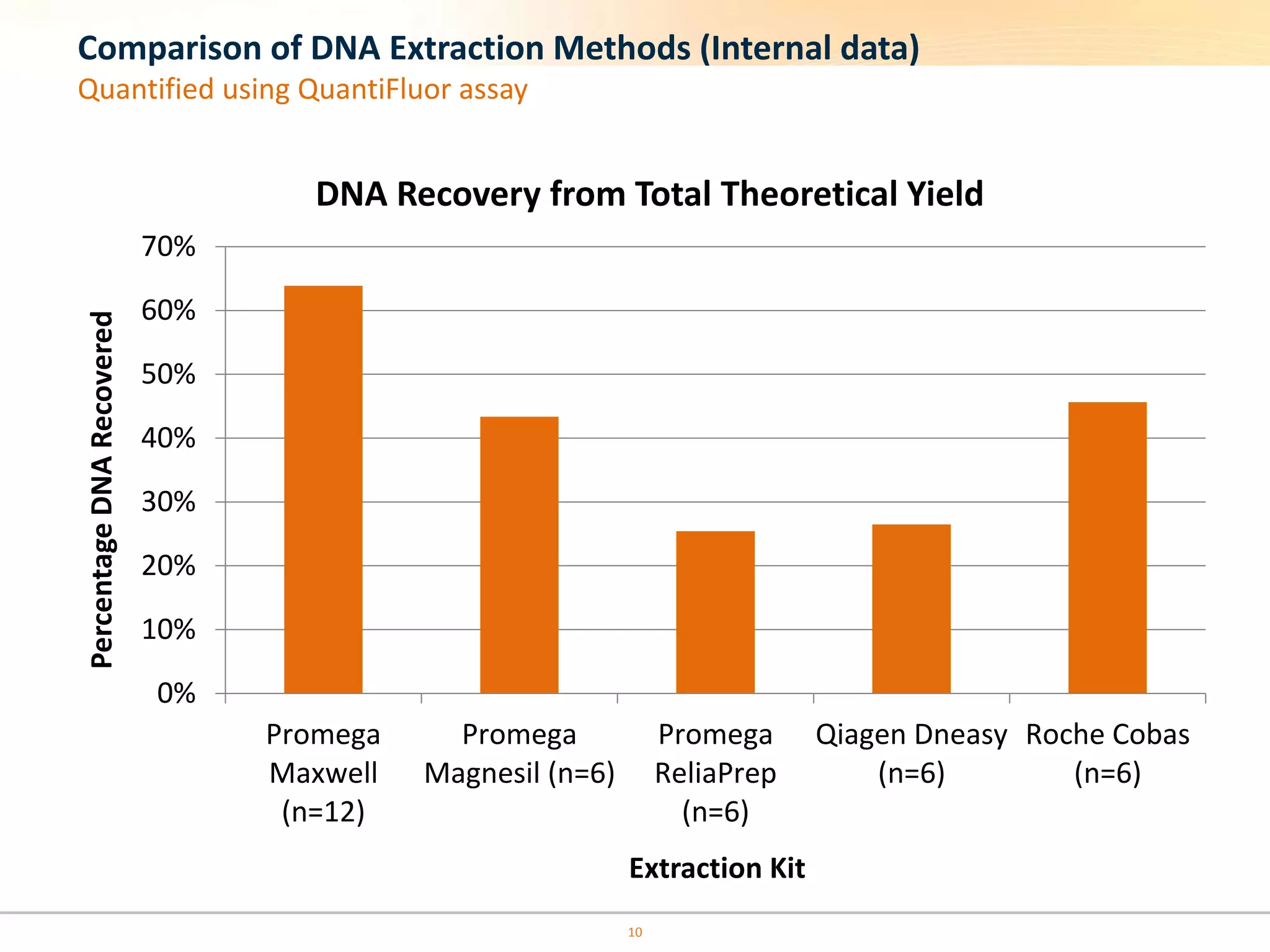
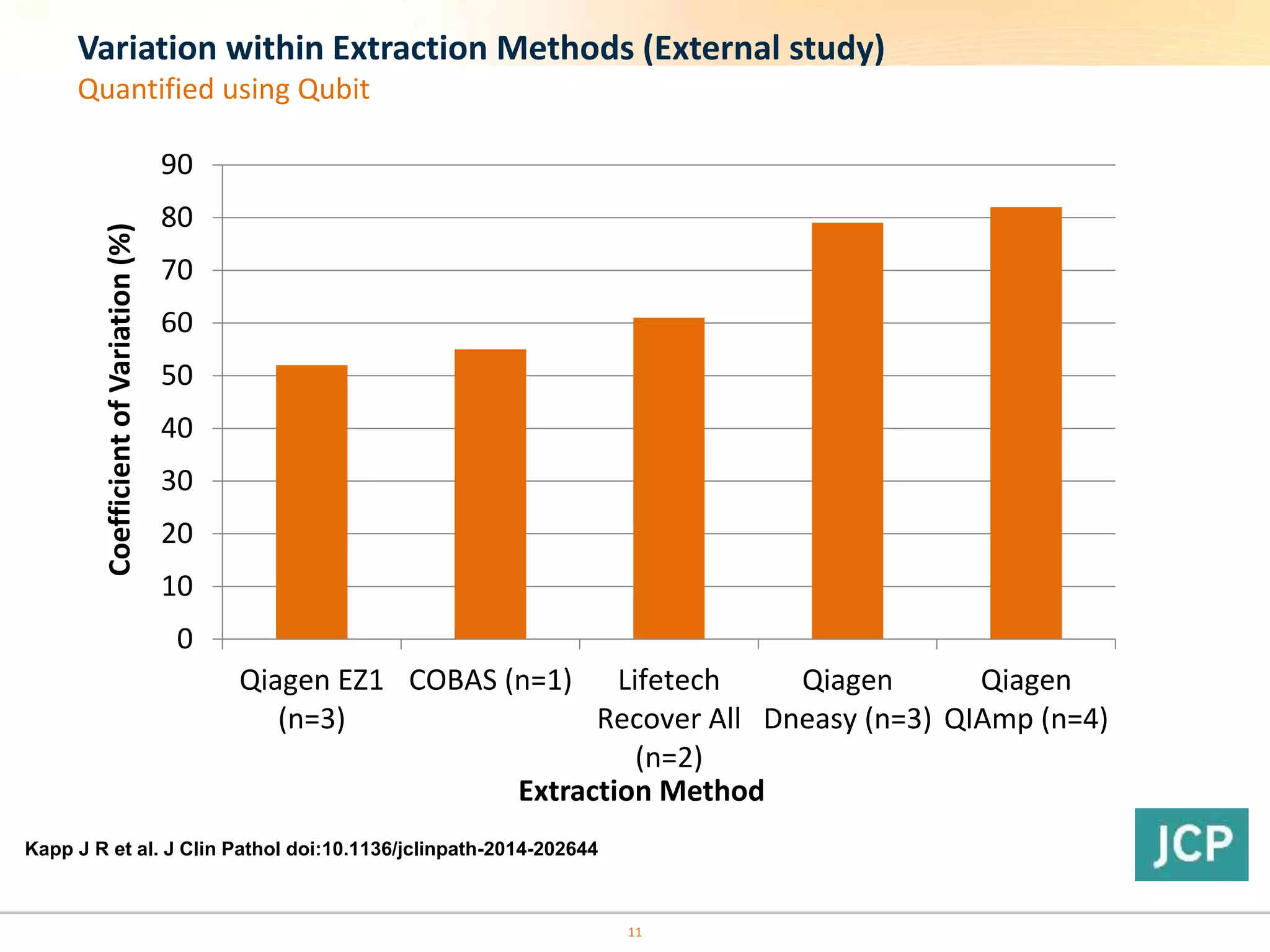
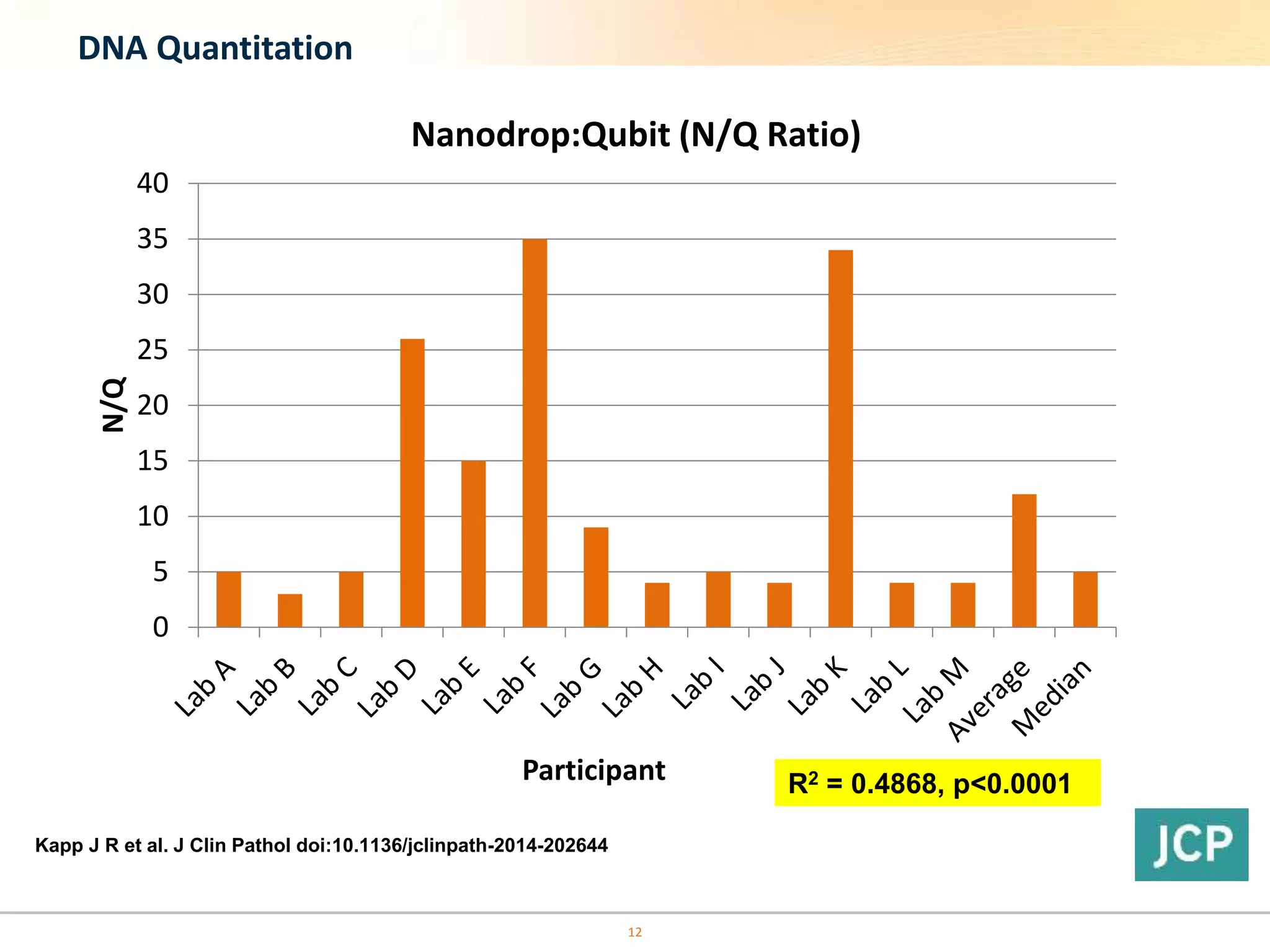
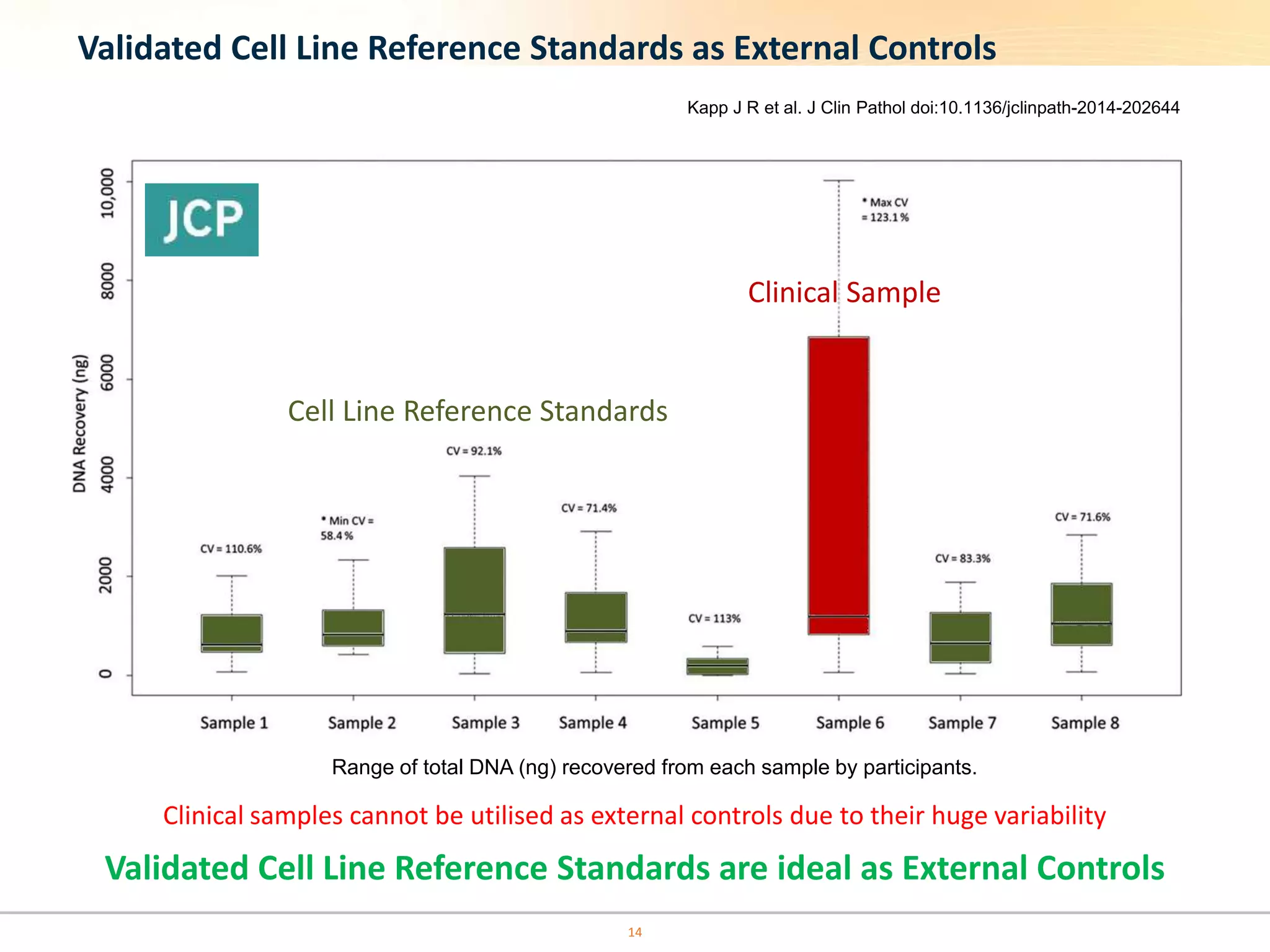
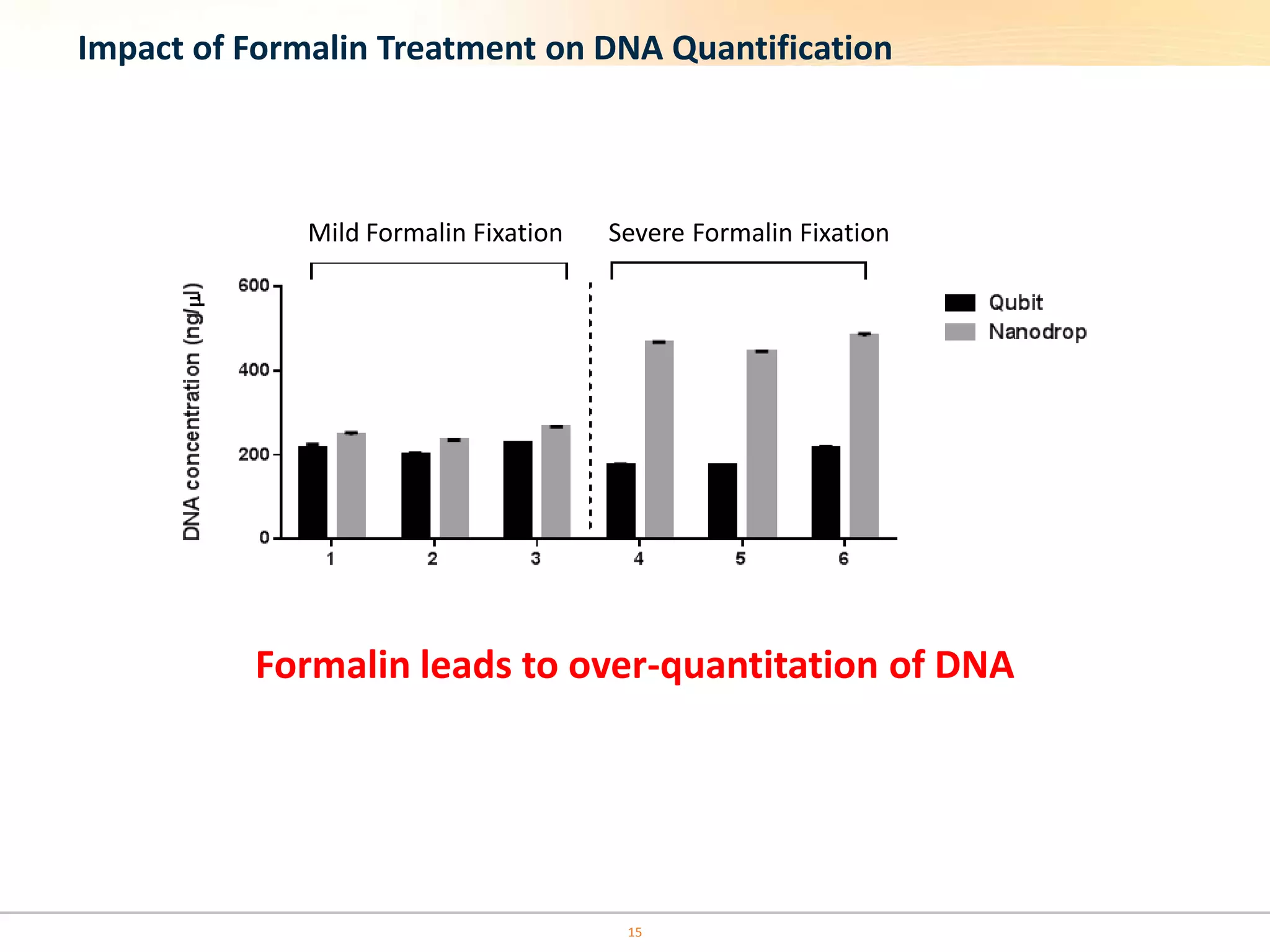
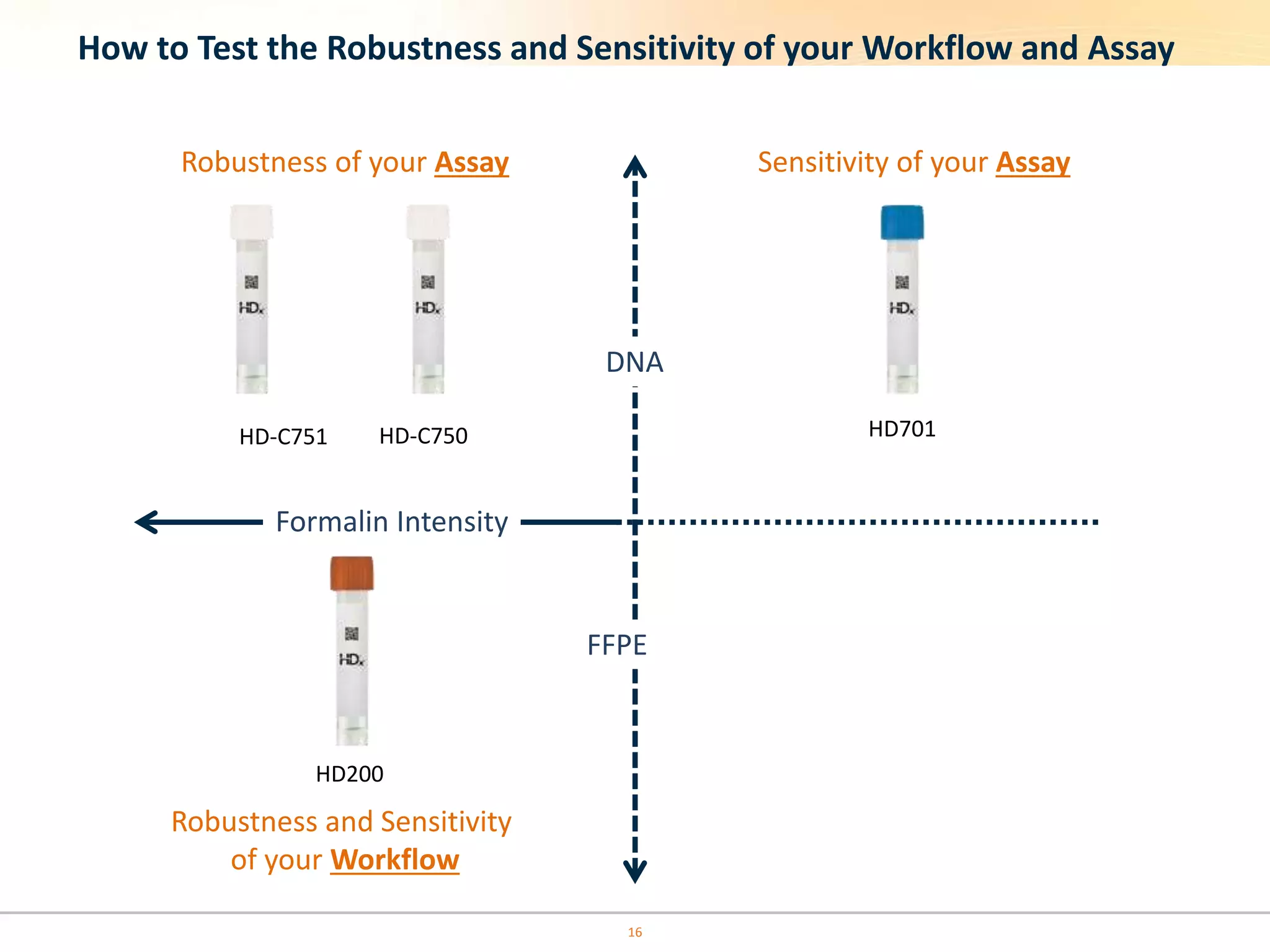
The document discusses the challenges of pre-PCR analytical variability in FFPE samples, emphasizing the impact of assay failures and the importance of monitoring methods. It highlights various extraction techniques and their differing efficiencies, along with the effect of formalin fixation on DNA quantification. The use of validated cell line reference standards as external controls is recommended to improve diagnostic accuracy and reliability.