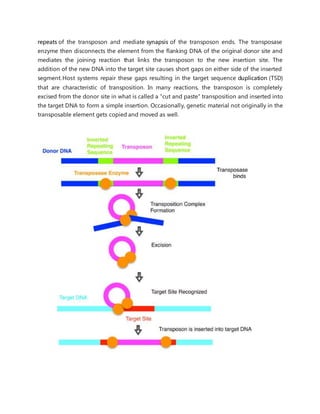
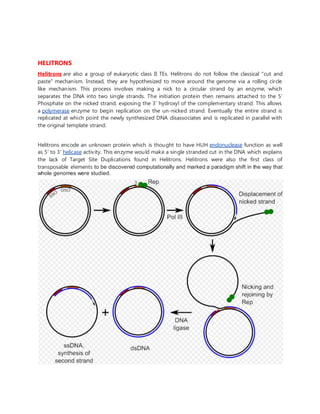
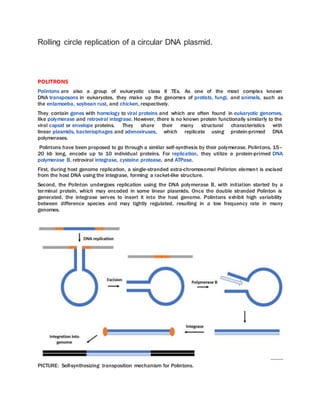
DNA transposons, also called jumping genes, can move to different locations within a genome. They move through a DNA intermediate rather than an RNA intermediate like retrotransposons. There are two main classes of DNA transposons - autonomous, which can move on their own, and non-autonomous, which require a transposase enzyme from another element to move. DNA transposons move around the genome via three main mechanisms: cut-and-paste transposition, rolling circle transposition (Helitrons), or self-synthesis (Polintons).