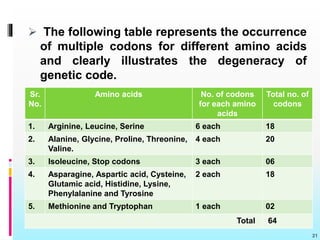
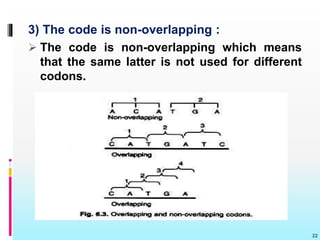
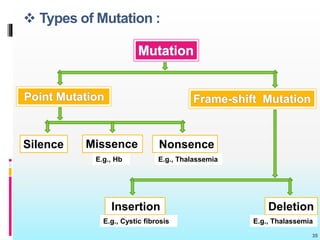
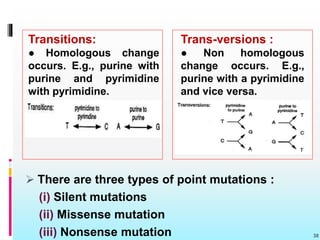
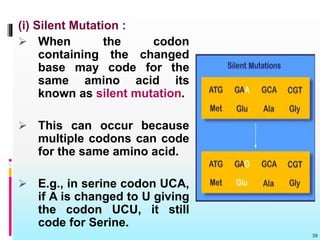
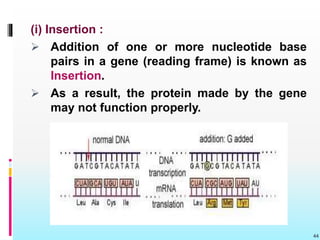
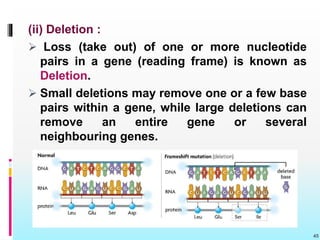
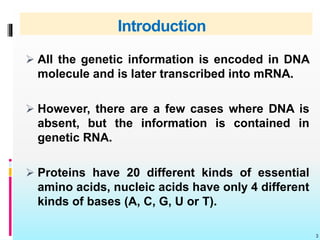
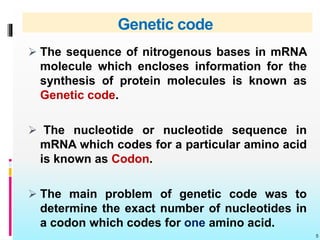
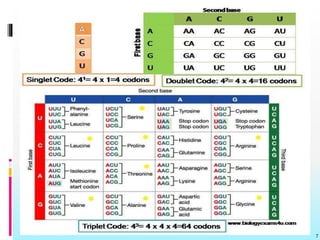
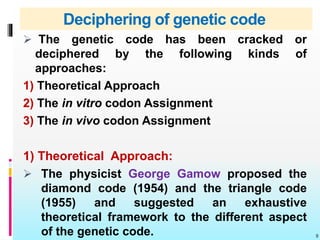
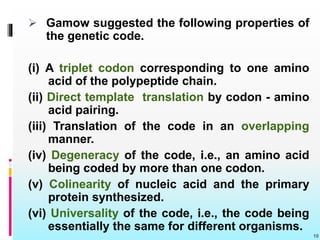
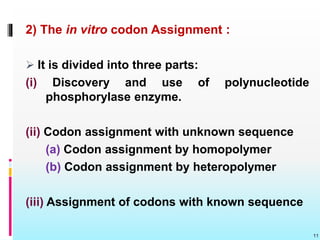
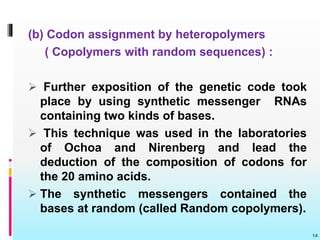
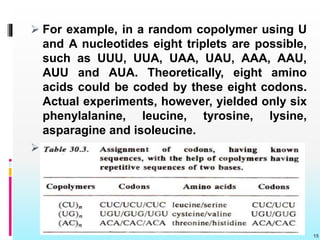
The document discusses the genetic code, detailing its components, deciphering methods, and properties. It explains the triplet nature of codons, their roles as initiation and termination signals, and the implications of gene mutations. It also highlights key experiments that led to the understanding of genetic coding in protein synthesis.





![(iii) Assignment of codons with known sequence:
[Use of trinucleotides or minimessengers in
filter binding (Ribosome – binding technique)]
Ribosome binding technique of Nirenberg and
Leder (1964) made use of the finding that
aminocyl-tRNA molecules specifically bind to
ribosome – mRNA complex.
This binding does not require the presence of a
long mRNA molecules; in fact, the association
of a trinucleotide or minimessenger with the
ribosome is sufficient to cause aminoacyl-tRNA
binding.
16](https://image.slidesharecdn.com/maitri216geneticcode-191231165536/85/Genetic-code-16-320.jpg)